Mapping MAGE-A4 expression in solid cancers for targeted therapies
- PMID: 40151801
- PMCID: PMC11947667
- DOI: 10.3389/fonc.2025.1484182
Mapping MAGE-A4 expression in solid cancers for targeted therapies
Abstract
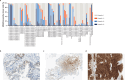
Melanoma-associated antigen A4 (MAGE-A4) is a promising target for anticancer therapy. However, limited contemporary data are available on the details of MAGE-A4 protein expression in different cancer types. In this study, the protein expression of MAGE-A4 is comprehensively studied in patients with unresectable and/or metastatic solid cancers to identify indications of the highest unmet medical need for anti-MAGE-A4 therapy. FFPE tumor sections from 200 patients, predominantly HLA-A*02:01 positive (n = 193), were examined using immunohistochemistry (IHC) to detect MAGE-A4 expression. The patient cohort comprised various cancer types to pinpoint differences in the prevalence and intensity of MAGE-A4 positivity. MAGE-A4 expression was observed in 35% (69 patients) of the overall cohort. Certain cancer types exhibited notably higher frequencies of MAGE-A4 positivity. Specifically, adenoid cystic carcinoma demonstrated the highest prevalence at 82%, followed by liposarcoma at 67%. Ovarian serous/high-grade carcinoma showed a 64% positivity rate, identical to that observed in squamous non-small cell lung cancer (NSCLC). Head and neck squamous cell carcinoma (HNSCC) presented a 60% prevalence, while esophageal cancer had a 54% prevalence of MAGE-A4 expression. These data highlight the variability of MAGE-A4 expression across different cancer types and underscore its relevance as a potential target of novel precision medicines. The significant presence of MAGE-A4 in specific cancers suggests potential for stratified therapeutic approaches and warrants further investigation into its role in oncogenesis and treatment response.
Keywords: biomarker; clinical trial; patient selection; target expression; translational analysis.
Copyright © 2025 Habigt, Rottey, Spanggaard, Lopez, Garralda, Calvo, Bechter, Desai, Galot, Gandhi, Heil, Rieder, Dimitrov, Quetglas, Heichinger, Keshelava and Roller.
Conflict of interest statement
CHa reports being an employee of Roche Diagnostics GmbH at the time of the study and is a shareholder of F. Hoffmann-La Roche Ltd. SR reports research grants for Bristol Myers-Squibb and F. Hoffmann-La Roche Ltd. and consulting fees for Ipsen, Pfizer, Astellas, F. Hoffmann-La Roche Ltd., Bristol Myers-Squibb, and Merck Sharp & Dohme. IS reports institutional grants from F. Hoffmann-La Roche Ltd., AstraZeneca, Genentech, Inc., Incyte, Merck Sharp & Dohme, Orion, Pfizer, Novartis, Genmab, Puma Biotechnology, and Bristol Myers-Squibb; honoraria for lectures from AstraZeneca; and support for attending meetings and/or travel from AstraZeneca, Incyte, and Merck/Pfizer. JL reports research funding for F. Hoffmann-La Roche Ltd., Genentech, Inc., Basilea, Astex, Merck Sharp & Dohme, Genmab, Janssen, Verastem, and Kazia; and advisory committees for Basilea, GlaxoSmithKline, Genmab, Servier, and F. Hoffmann-La Roche Ltd. EG reports consultancy for F. Hoffmann-La Roche Ltd., Ellipses Pharma, Boehringer Ingelheim, Janssen Global Services, Seattle Genetics, Thermo Fisher, MAB Discovery, Anaveon, Hengrui, Sanofi, Incyte, Medscape, Pfizer, and Amgen; research funding for Novartis, F. Hoffmann-La Roche Ltd., Thermo Fisher, AstraZeneca, Taiho, BeiGene, and Janssen; speaker’s bureaus for Merck Sharp & Dohme, F. Hoffmann-La Roche Ltd., Thermo Fisher, Novartis, and SeaGen; and stocks with 1TRIALSP. EC reports support on advisory boards for Adcendo, Amunix, Anaveon, AstraZeneca, Bristol Myers-Squibb, Chugai, Diaccurate, Elevation Oncology, Ellipses Pharmacy, Genmab, Janssen, MonTa, Merck Sharp & Dohme, Nanobiotix, Nouscom, Novartis, Servier, Syneos Health, T-knife, and TargIumme; invited speaker for OncoDNA, PharmaMar, and F. Hoffmann-La Roche Ltd./Genentech, Inc.; ownership interest for Oncoart Associated and START; part of full-time employment for HM Hospitales Group and START Madrid – COICC; board director member for PharmaMar; steering committee member for BeiGene, Merus, Novartis, and Sanofi; non-financial advisory role for CRIS Cancer Foundation; non-financial advisory role for PsiOxus; non-financial member of ASCO, EORTC, ESMO, SEOM, and Non-for-profit Foundation PharMa; non-financial president and co-founder of INTHEOS; and non-financial chair of Independent Data Monitoring Committee for EORTC IDMC. OB reports consultancy for Merck Sharp & Dohme, Bristol Myers-Squibb, Ultimovacs, Sanofi, and Sun Pharmaceutical Industries Ltd. JD reports consulting or advisory roles for BeiGene, Pierre Fabre, Bayer, GlaxoSmithKline, Merck KGaA, Boehringer Ingelheim, F. Hoffmann-La Roche Ltd./Genentech, Inc., Daiichi Sankyo Europe GmbH, Novartis, Pfizer, Ellipses Pharma, Axelia Oncology, Incyte, and Amgen; and research funding for F. Hoffmann-La Roche Ltd., GlaxoSmithKline, Novartis, BeiGene, Bristol Myers-Squibb, AstraZeneca/MedImmune, and Amgen. RG reports speaker’s bureau for Bristol Myers-Squibb and travel expenses for Merck and Merck Sharp & Dohme. LG reports employment and equity from NextPoint Therapeutics; advisory committees from Accurius; and board membership from BrightPeak Therapeutics and Neximmune. FH and NR report being employees of Roche Diagnostics GmbH at the time of the study. ID reports being an employee of Roche Tissue Diagnostics at the time of the study. IQ reports being an employee of F. Hoffmann-La Roche Ltd. at the time of the study and is a shareholder of F. Hoffmann-La Roche Ltd. CHe and NK report being employees of F. Hoffmann-La Roche Ltd. at the time of the study. AR reports being an employee of F. Hoffmann-La Roche at the time of the study and is a shareholder of F. Hoffmann-La Roche Ltd. All authors report medical writing support on the present manuscript. The authors declare that this study received funding from F. Hoffmann-La Roche Ltd. The funder had the following involvement in the study: study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

