Protocol for spatial metabolomics and isotope tracing in the mouse brain
- PMID: 40153438
- PMCID: PMC11995059
- DOI: 10.1016/j.xpro.2025.103716
Protocol for spatial metabolomics and isotope tracing in the mouse brain
Abstract
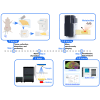
Mass spectrometry imaging enables high-resolution spatial chemical mapping, yet its application for dynamic analysis with tracers poses challenges. Here, we present a protocol for spatial metabolomics and isotope tracing in the mouse brain. We describe steps for tracer administration, tissue collection, and cryosectioning. We then detail procedures for matrix application, ion identification, and data analysis. This protocol delivers high-quality spatial metabolomics data and is well suited for region-specific tracing analysis in the brain.
Keywords: mass spectrometry; metabolism; neuroscience.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.Y.-S., M.A.H., and M.N.B. were employed by Shimadzu Scientific Instruments, Inc., and S.Y. was employed by Shimadzu Corporation during the development of the protocol and/or preparation of the manuscript.
Figures




References
-
- Marin-Valencia I., Kocabas A., Rodriguez-Navas C., Miloushev V.Z., González-Rodríguez M., Lees H., Henry K.E., Vaynshteyn J., Longo V., Deh K., et al. Imaging brain glucose metabolism in vivo reveals propionate as a major anaplerotic substrate in pyruvate dehydrogenase deficiency. Cell Metab. 2024;36:1394–1410.e12. doi: 10.1016/j.cmet.2024.05.002. - DOI - PMC - PubMed
-
- Heinrich P., Kohler C., Ellmann L., Kuerner P., Spang R., Oefner P.J., Dettmer K. Correcting for natural isotope abundance and tracer impurity in MS-MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-36293-4. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

