Ecotin protects Salmonella Typhimurium against the microbicidal activity of host proteases
- PMID: 40153455
- PMCID: PMC11977995
- DOI: 10.1371/journal.ppat.1013013
Ecotin protects Salmonella Typhimurium against the microbicidal activity of host proteases
Abstract
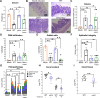
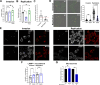
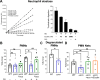
Salmonella enterica serovar Typhimurium causes acute diarrhea upon oral infection in humans. The harsh and proteolytic environment found in the gastrointestinal tract is the first obstacle that these bacteria face after infection. However, the mechanisms that allow Salmonella to survive the hostile conditions of the gut are poorly understood. The ecotin gene is found in an extensive range of known phyla of bacteria and it encodes a protein that has been shown to inhibit serine proteases. Thus, in the present work we studied the role of ecotin of Salmonella Typhimurium in host-pathogen interactions. We found that the Salmonella Typhimurium ∆ecotin strain exhibited lower inflammation in a murine model of Salmonella induced colitis. The ∆ecotin mutant was more susceptible to the action of pancreatin and purified pancreatic elastase. In addition, the lack of ecotin led to impaired adhesion to Caco-2 and HT-29 cell lines, related to the proteolytic activity of brush border enzymes. Besides, ∆ecotin showed higher susceptibility to lysosomal proteolytic content and intracellular replication defects in macrophages. In addition, we found Ecotin to have a crucial role in Salmonella against the microbicidal action of granule contents and neutrophil extracellular traps released from human polymorphonuclear leukocytes. Thus, the work presented here highlights the importance of ecotin in Salmonella as countermeasures against the host proteolytic defense system.
Copyright: © 2025 Saposnik et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal's policy and JC, KP and LC have the following competing interests: JC, KP and LC are inventors of a patent related to Ecotin. The patent “Immunomodulating and immunostimulating polypeptides for drug-delivery” was filled on February 8, 2019 by the authors’ National Research Council (CONICET) and the University of San Martin (UNSAM) in the United States patent and trademark Office (US2021009371), the European patent office (PCT/IB2019/051026, EP3749364), the China National intellectual property administration (CN111936166), and the National Institute of Intellectual Property from Argentina (AR114683). The filing of the patent did not have any role in experimental design, data collection and analysis, decision to publish, or preparation of this manuscript.
Figures


Update of
-
Ecotin protects Salmonella Typhimurium against the microbicidal activity of host proteases.bioRxiv [Preprint]. 2024 May 16:2024.05.15.594389. doi: 10.1101/2024.05.15.594389. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Mar 28;21(3):e1013013. doi: 10.1371/journal.ppat.1013013. PMID: 38798423 Free PMC article. Updated. Preprint.

