A novel human organoid model system reveals requirement of TCF4 for oligodendroglial differentiation
- PMID: 40155049
- PMCID: PMC11953572
- DOI: 10.26508/lsa.202403102
A novel human organoid model system reveals requirement of TCF4 for oligodendroglial differentiation
Abstract
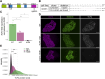
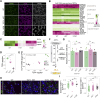
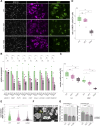
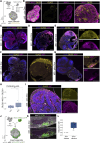
Heterozygous mutations of TCF4 in humans cause Pitt-Hopkins syndrome, a neurodevelopmental disease associated with intellectual disability and brain malformations. Although most studies focus on the role of TCF4 in neural stem cells and neurons, we here set out to assess the implication of TCF4 for oligodendroglial differentiation. We discovered that both monoallelic and biallelic mutations in TCF4 result in a diminished capacity to differentiate human neural progenitor cells toward myelinating oligodendrocytes through the forced expression of the transcription factors SOX10, OLIG2, and NKX6.2. Using this experimental strategy, we established a novel organoid model, which generates oligodendroglial cells within a human neurogenic tissue-like context. Also, here we found a reduced ability of TCF4 heterozygous cells to differentiate toward oligodendroglial cells. In sum, we establish a role of human TCF4 in oligodendrocyte differentiation and provide a model system, which allows to dissect the disease etiology in a human tissue-like context.
© 2025 Furlanetto et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Avery S, Hirst AJ, Baker D, Lim CY, Alagaratnam S, Skotheim RI, Lothe RA, Pera MF, Colman A, Robson P, et al. (2013) BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Rep 1: 379–386. 10.1016/j.stemcr.2013.10.005 - DOI - PMC - PubMed
-
- Bohlen JF, Cleary CM, Das D, Sripathy SR, Sadowski N, Shim G, Kenney RF, Buchler IP, Banerji T, Scanlan TS, et al. (2023) Promyelinating drugs promote functional recovery in an autism spectrum disorder mouse model of Pitt-Hopkins syndrome. Brain 146: 3331–3346. 10.1093/brain/awad057 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
