Identification of aberrant interferon-stimulated gene associated host responses potentially linked to poor prognosis in COVID-19 during the Omicron wave
- PMID: 40155905
- PMCID: PMC11954226
- DOI: 10.1186/s12985-025-02696-9
Identification of aberrant interferon-stimulated gene associated host responses potentially linked to poor prognosis in COVID-19 during the Omicron wave
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron has demonstrated decreased pathogenicity, yet a few individuals suffer severe pneumonia from coronavirus disease 2019 (COVID-19) infection; the underlying mechanisms are unknown.
Methods: The present work investigated the role of Interferon-stimulated genes (ISGs) in the occurrence and progression of severe Omicron infection. The expression and dynamic changes of ISGs were assessed using quantitative real-time polymerase chain reaction (qRT-PCR), and the anti-ISG15 autoantibody in infected patients was detected by ELISA. Moreover, we evaluated the correlation of ISGs with disease severity and outcomes by comparing expression of ISGs among each group.
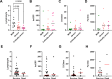
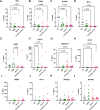
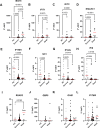
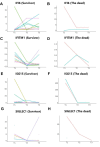
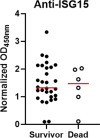
Results: Decreased expression of several ISGs such as IFI6 are potentially linked to increased severity or poor outcomes of Omicron infection. Longitudinal data also demonstrates that the dynamic variation of IFI6 in the Omicron infection phase may be linked to the prognosis of the disease. The increase of anti-ISG15 autoantibody potentially links to the disease progression and poor outcome of patients with high level of ISG15 expression.
Conclusions: These findings define aberrant Interferon-stimulated gene associated host responses and reveal potential mechanisms and therapeutic targets for Omicron or other viral infection.
Keywords: Antiviral restriction factors; COVID-19; ISG; Interferons; SARS-CoV-2 Omicron variant.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), was approved by the Institutional Review Committee of Peking Union Medical College Hospital (approval number: I-23PJ292). Written informed consent was obtained from all participants. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
-
Restriction of SARS-CoV-2 replication by receptor transporter protein 4 (RTP4).mBio. 2023 Aug 31;14(4):e0109023. doi: 10.1128/mbio.01090-23. Epub 2023 Jun 29. mBio. 2023. PMID: 37382452 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

