Benchmarking spatial transcriptomics technologies with the multi-sample SpatialBenchVisium dataset
- PMID: 40156041
- PMCID: PMC11954323
- DOI: 10.1186/s13059-025-03543-4
Benchmarking spatial transcriptomics technologies with the multi-sample SpatialBenchVisium dataset
Abstract
Background: Spatial transcriptomics allows gene expression to be measured within complex tissue contexts. Among the array of spatial capture technologies available is 10x Genomics' Visium platform, a popular method which enables transcriptome-wide profiling of tissue sections. Visium offers a range of sample handling and library construction methods which introduces a need for benchmarking to compare data quality and assess how well the technology can recover expected tissue features and biological signatures.
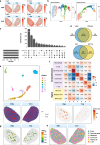
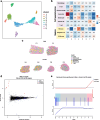
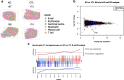
Results: Here we present SpatialBenchVisium, a unique reference dataset generated from spleen tissue of mice responding to malaria infection spanning several tissue preparation protocols (both fresh frozen and FFPE, with either manual or CytAssist tissue placement). We note better quality control metrics in reference samples prepared using probe-based capture methods, particularly those processed with CytAssist, validating the improvement in data quality produced with the platform. Our analysis of replicate samples extends to explore spatially variable gene detection, the outcomes of clustering and cell deconvolution using matched single-cell RNA-sequencing data and publicly available reference data to identify cell types and tissue regions expected in the spleen. Multi-sample differential expression analysis recovered known gene signatures related to biological sex or gene knockout.
Keywords: 10x Visium; Benchmarking; Differential expression; Multi-sample analysis; Spatial transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experiments were performed with ethics approval from the Animal Ethics Committee of the Walter and Eliza Hall Institute of Medical Research. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Marx V. Method of the year: spatially resolved transcriptomics. Nat Methods. 2021;1(18):9–14. 10.1038/s41592-020-01033-y. - PubMed
-
- He S, Bhatt R, Brown C, Brown EA, Buhr DL, Chantranuvatana K, et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol. 2022;40:1794–806. 10.1038/s41587-022-01483-z. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

