Gastroesophageal circulating tumor cell crosstalk with peripheral immune system guides CTC survival and proliferation
- PMID: 40157906
- PMCID: PMC11954855
- DOI: 10.1038/s41419-025-07530-2
Gastroesophageal circulating tumor cell crosstalk with peripheral immune system guides CTC survival and proliferation
Abstract
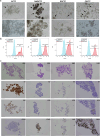
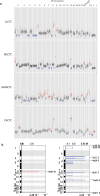
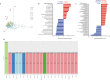
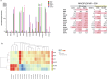
Tumor dissemination is a key event in tumor progression. During this event, a main role is played by circulating tumor cells (CTCs), immune cells, and their interaction. How the immune system supports the survival and proliferation of CTCs is not fully elucidated. In this study we established an in-vitro co-culture system consisting of immune cells and CTCs from the same patient, which increased the success rate in the establishment of CTC-derived long-term cell cultures. In this system, we characterized the immune cells of successful co-cultures and the signals they exchange with cancer cells, including cytokines and extracellular vesicle (EV) content. Using this protocol, we stabilized four CTC-derived cell lines from patients with metastatic gastroesophageal cancer, which were cultured for over a year and characterized from a genetic and molecular point of view. The four cell lines harbor shared chromosomal aberrations including the amplification at 8q24.21 containing MYC and deletion 9p21.3 containing CDKN2A/B and the IFN type I cluster. The transcriptomic profile of CTC cell lines is distinct from primary tumors, and we detected the activation of E2F, G2M and MYC pathways and the downregulation of interferon response pathway. Each cell line shows a degree of invasiveness in zebrafish in-vivo, and the most invasive ones share the same mutation in RAB14 gene. In addition, the four cell lines secrete cell-line specific EVs containing microRNAs that target YAP, BRG1-AKT1, TCF8-HDAC pathways. Overall, we highlight how the immune system plays a key role in the proliferation of CTCs through EV signaling, and how CTC cell line genomic and transcriptomic alterations make these cells less visible from the immune system and likely responsible for the survival advantage in sites distant from the microenvironment of origin.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Research Ethics Committee from IRCCS Istituto Romagnolo per lo Studio dei Tumori (IRST) “Dino Amadori” (Italy) (protocol code, IRSTB063, date of approval 11 May 2016).
Figures


References
-
- Morgan E, Soerjomataram I, Rumgay H, Coleman HG, Thrift AP, Vignat J, et al. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology. 2022;163:649–658.e2. - PubMed
-
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

