Advancing CRISPR genome editing into gene therapy clinical trials: progress and future prospects
- PMID: 40160040
- PMCID: PMC12094669
- DOI: 10.1017/erm.2025.10
Advancing CRISPR genome editing into gene therapy clinical trials: progress and future prospects
Abstract
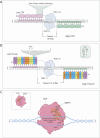
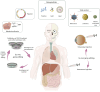
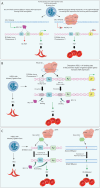
Genome editing has recently evolved from a theoretical concept to a powerful and versatile set of tools. The discovery and implementation of CRISPR-Cas9 technology have propelled the field further into a new era. This RNA-guided system allows for specific modification of target genes, offering high accuracy and efficiency. Encouraging results are being announced in clinical trials employed in conditions like sickle cell disease (SCD) and transfusion-dependent beta-thalassaemia (TDT). The path finally led the way to the recent FDA approval of the first gene therapy drug utilising the CRISPR/Cas9 system to edit autologous CD34+ haematopoietic stem cells in SCD patients (Casgevy). Ongoing research explores the potential of CRISPR technology for cancer therapies, HIV treatment and other complex diseases. Despite its remarkable potential, CRISPR technology faces challenges such as off-target effects, suboptimal delivery systems, long-term safety concerns, scalability, ethical dilemmas and potential repercussions of genetic alterations, particularly in the case of germline editing. Here, we examine the transformative role of CRISPR technologies, including base editing and prime editing approaches, in modifying the genetic and epigenetic codes in the human genome and provide a comprehensive focus, particularly on relevant clinical applications, to unlock the full potential and challenges of gene editing.
Keywords: CRISPR/Cas; gene editing; gene therapy; genetic diseases; medical genetics.
Conflict of interest statement
The authors declare none.
Figures






References
-
- Shamshirgaran Y, Liu J, Sumer H, Verma PJand Taheri-Ghahfarokhi A (2022) Tools for efficient genome editing; ZFN, TALEN, and CRISPR. Methods in Molecular Biology 2495,29–46. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

