Identification of biomarkers associated with programmed cell death in liver ischemia-reperfusion injury: insights from machine learning frameworks and molecular docking in multiple cohorts
- PMID: 40160318
- PMCID: PMC11949969
- DOI: 10.3389/fmed.2025.1501467
Identification of biomarkers associated with programmed cell death in liver ischemia-reperfusion injury: insights from machine learning frameworks and molecular docking in multiple cohorts
Abstract
Introduction: Liver ischemia-reperfusion injury (LIRI) is a major reason for liver injury that occurs during surgical procedures such as hepatectomy and liver transplantation and is a major cause of graft dysfunction after transplantation. Programmed cell death (PCD) has been found to correlate with the degree of LIRI injury and plays an important role in the treatment of LIRI. We aim to comprehensively explore the expression patterns and mechanism of action of PCD-related genes in LIRI and to find novel molecular targets for early prevention and treatment of LIRI.
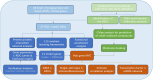
Methods: We first compared the expression profiles, immune profiles, and biological function profiles of LIRI and control samples. Then, the potential mechanisms of PCD-related differentially expressed genes in LIRI were explored by functional enrichment analysis. The hub genes for LIRI were further screened by applying multiple machine learning methods and Cytoscape. GSEA, GSVA, immune correlation analysis, transcription factor prediction, ceRNA network analysis, and single-cell analysis further revealed the mechanisms and regulatory network of the hub gene in LIRI. Finally, potential therapeutic agents for LIRI were explored based on the CMap database and molecular docking technology.
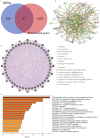
Results: Forty-seven differentially expressed genes associated with PCD were identified in LIRI, and functional enrichment analysis showed that they were involved in the regulation of the TNF signaling pathway as well as the regulation of hydrolase activity. By utilizing machine learning methods, 11 model genes were identified. ROC curves and confusion matrix from the six cohorts illustrate the superior diagnostic value of our model. MYC was identified as a hub PCD-related target in LIRI by Cytoscape. Finally, BMS-536924 and PF-431396 were identified as potential therapeutic agents for LIRI.
Conclusion: This study comprehensively characterizes PCD in LIRI and identifies one core molecule, providing a new strategy for early prevention and treatment of LIRI.
Keywords: biomarkers; liver ischemia–reperfusion injury; machine learning; molecular docking; programmed cell death.
Copyright © 2025 Liu, Jin, Lv, Yang, Li, Zhang, Zhong and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

