Out-of-Africa migration and clonal expansion of a recombinant Epstein-Barr virus drives frequent nasopharyngeal carcinoma in southern China
- PMID: 40160679
- PMCID: PMC11954593
- DOI: 10.1093/nsr/nwae438
Out-of-Africa migration and clonal expansion of a recombinant Epstein-Barr virus drives frequent nasopharyngeal carcinoma in southern China
Abstract
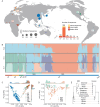
While Epstein-Barr virus (EBV) infection is ubiquitous globally, a high-risk EBV subtype associated with the extremely high incidence of nasopharyngeal carcinoma (NPC) was found in southern China, but the evolution history of EBV in humans and the origin of this high-risk subtype remains enigmatic. By obtaining one of the largest datasets of EBV genomes across the world, we found that EBV had an evolutionary history matching the out-of-Africa migration of humans. Within the high-risk subtype from southern China, we identified a rapidly expanding clonal strain originating from a recombination event between EBV strains from northern and southern Chinese around 4000 years ago, followed by strong Darwinian evolution with a fitness advantage of 4%. The clonal strain has almost doubled the risk for NPC compared to the high-risk subtype and explained around 66% of the NPCs, representing the highest risk factor for NPC identified so far. Taken together, we unraveled a strong co-evolution history between EBV and humans where human migration and admixture triggered subsequent recombination and expansion of a highly advantageous EBV strain, leading to a cancer epidemic in southern China.
Keywords: Epstein-Barr virus; Nasopharyngeal carcinoma; co-evolution, adaptation; out-of-Africa migration; recombination.
© The Author(s) 2024. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures



References
LinkOut - more resources
Full Text Sources
