This is a preprint.
Stability and DNA Methyltransferase Activity of DNMT3A are Maintained by Ubiquitin-Specific Peptidase 11 (USP11) and Sumoylation Countering Degradation
- PMID: 40161590
- PMCID: PMC11952362
- DOI: 10.1101/2025.03.05.641683
Stability and DNA Methyltransferase Activity of DNMT3A are Maintained by Ubiquitin-Specific Peptidase 11 (USP11) and Sumoylation Countering Degradation
Abstract
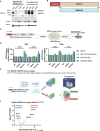
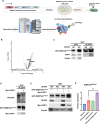
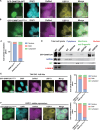
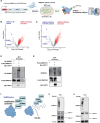
DNA methyltransferase 3A (DNMT3A) plays crucial roles in mammalian development and hematopoiesis. DNMT3A protein instability is associated with blood diseases such as myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), as well as Tatton-Brown-Rahman syndrome, an overgrowth disorder. We found that certain unstable DNMT3A mutations cause DNMT3A localization changes, resulting in loss of function. This mislocalization is partially rescued by E1 enzyme inhibition or stable USP11 expression, as DNMT3A stability is maintained by deubiquitinating enzyme USP11 countering degradation by CUL4-DCAF8 E3 ligase. We also found that USP11 enhances DNMT3A SUMOylation by promoting the interaction between DNMT3A and SUMO E3 ligases. DNMT3A SUMOylation also is essential to maintain DNMT3A protein stability. Furthermore, we found USP11 enhances the binding of DNMT3A to the polycomb complex and maintains DNMT3A DNA methyltransferase (MTase) activity. This study uncovers the mechanism for DNMT3A protein turnover through USP11, which is essential to DNMT3A function, and may be a therapeutic approach for diseases caused by DNMT3A protein instability.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
