This is a preprint.
Rapidly evolved genomic regions shape individual language abilities in present-day humans
- PMID: 40161630
- PMCID: PMC11952349
- DOI: 10.1101/2025.03.07.641231
Rapidly evolved genomic regions shape individual language abilities in present-day humans
Abstract
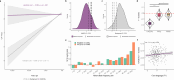
1Minor genetic changes have produced profound differences in cognitive abilities between humans and our closest relatives, particularly in language. Despite decades of research, ranging from single-gene studies to broader evolutionary analyses[1, 2, 3, 4, 5], key questions about the genomic foundations of human language have persisted, including which sequences are involved, how they evolved, and whether similar changes occur in other vocal learning species. Here we provide the first evidence directly linking rapidly evolved genomic regions to language abilities in contemporary humans. Through extensive analysis of 65 million years of evolutionary events in over 30,000 individuals, we demonstrate that Human Ancestor Quickly Evolved Regions (HAQERs)[5] - sequences that rapidly accumulated mutations after the human-chimpanzee split - specifically influence language but not general cognition. These regions evolved to shape language development by altering binding of Forkhead domain transcription factors, including FOXP2. Strikingly, language-associated HAQER variants show higher prevalence in Neanderthals than modern humans, have been stable throughout recent human history, and show evidence of convergent evolution across other mammalian vocal learners. An unexpected pattern of balancing selection acting on these apparently beneficial alleles is explained by their pleiotropic effects on prenatal brain development contributing to birth complications, reflecting an evolutionary trade-off between language capability and reproductive fitness. By developing the Evolution Stratified-Polygenic Score analysis, we show that language capabilities likely emerged before the human-Neanderthal split - far earlier than previously thought[3, 6, 7]. Our findings establish the first direct link between ancient genomic divergence and present-day variation in language abilities, while revealing how evolutionary constraints continue to shape human cognitive development.
Figures




References
-
- Lai C. S. L., Fisher S. E., Hurst J. A., Vargha-Khadem F., and Monaco A. P., “A forkhead-domain gene is mutated in a severe speech and language disorder,” Nature, vol. 413, pp. 519–523, 10 2001. - PubMed
-
- Vernes S. C., Newbury D. F., Abrahams B. S., Winchester L., Nicod J., Groszer M., Alarcón M., Oliver P. L., Davies K. E., Geschwind D. H., Monaco A. P., and Fisher S. E., “A functional genetic link between distinct developmental language disorders,” New England Journal of Medicine, vol. 359, pp. 2337–2345, 11 2008. - PMC - PubMed
-
- Mangan R. J., Alsina F. C., Mosti F., Sotelo-Fonseca J. E., Snellings D. A., Au E. H., Carvalho J., Sathyan L., Johnson G. D., Reddy T. E., Silver D. L., and Lowe C. B., “Adaptive sequence divergence forged new neurodevelopmental enhancers in humans.,” Cell, vol. 185, pp. 4587–4603.e23, 11 2022. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
