This is a preprint.
Sequencing-free whole genome spatial transcriptomics at molecular resolution in intact tissue
- PMID: 40161724
- PMCID: PMC11952344
- DOI: 10.1101/2025.03.06.641951
Sequencing-free whole genome spatial transcriptomics at molecular resolution in intact tissue
Update in
-
Sequencing-free whole-genome spatial transcriptomics at single-molecule resolution.Cell. 2025 Nov 26;188(24):6953-6970.e12. doi: 10.1016/j.cell.2025.09.006. Epub 2025 Oct 1. Cell. 2025. PMID: 41038164 Free PMC article.
Abstract
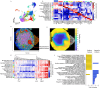
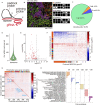
Recent breakthroughs in spatial transcriptomics technologies have enhanced our understanding of diverse cellular identities, compositions, interactions, spatial organizations, and functions. Yet existing spatial transcriptomics tools are still limited in either transcriptomic coverage or spatial resolution. Leading spatial-capture or spatial-tagging transcriptomics techniques that rely on in-vitro sequencing offer whole-transcriptome coverage, in principle, but at the cost of lower spatial resolution compared to image-based techniques. In contrast, high-performance image-based spatial transcriptomics techniques, which rely on in situ hybridization or in situ sequencing, achieve single-molecule spatial resolution and retain sub-cellular morphologies, but are limited by probe libraries that target only a subset of the transcriptome, typically covering several hundred to a few thousand transcript species. Together, these limitations hinder unbiased, hypothesis-free transcriptomic analyses at high spatial resolution. Here we develop a new image-based spatial transcriptomics technology termed Reverse-padlock Amplicon Encoding FISH (RAEFISH) with whole-genome level coverage while retaining single-molecule spatial resolution in intact tissues. We demonstrate image-based spatial transcriptomics targeting 23,000 human transcript species or 22,000 mouse transcript species, including nearly the entire protein-coding transcriptome and several thousand long-noncoding RNAs, in single cells in cultures and in tissue sections. Our analyses reveal differential subcellular localizations of diverse transcripts, cell-type-specific and cell-type-invariant tissue zonation dependent transcriptome, and gene expression programs underlying preferential cell-cell interactions. Finally, we further develop our technology for direct spatial readout of gRNAs in an image-based high-content CRISPR screen. Overall, these developments provide the research community with a broadly applicable technology that enables high-coverage, high-resolution spatial profiling of both long and short, native and engineered RNA species in many biomedical contexts.
Conflict of interest statement
Declaration of Interests S.W. and Y.Cheng are inventors on a patent application filed by Yale University related to RAEFISH and Perturb-RAEFISH reported in this manuscript. S.W. is one of the inventors on a patent applied for by Harvard University related to MERFISH.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
