This is a preprint.
Deciphering lung adenocarcinoma evolution and the role of LINE-1 retrotransposition
- PMID: 40161734
- PMCID: PMC11952568
- DOI: 10.1101/2025.03.14.643063
Deciphering lung adenocarcinoma evolution and the role of LINE-1 retrotransposition
Abstract
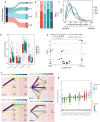
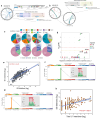
Understanding lung cancer evolution can identify tools for intercepting its growth. In a landscape analysis of 1024 lung adenocarcinomas (LUAD) with deep whole-genome sequencing integrated with multiomic data, we identified 542 LUAD that displayed diverse clonal architecture. In this group, we observed an interplay between mobile elements, endogenous and exogenous mutational processes, distinct driver genes, and epidemiological features. Our results revealed divergent evolutionary trajectories based on tobacco smoking exposure, ancestry, and sex. LUAD from smokers showed an abundance of tobacco-related C:G>A:T driver mutations in KRAS plus short subclonal diversification. LUAD in never smokers showed early occurrence of copy number alterations and EGFR mutations associated with SBS5 and SBS40a mutational signatures. Tumors harboring EGFR mutations exhibited long latency, particularly in females of European-ancestry (EU_N). In EU_N, EGFR mutations preceded the occurrence of other driver genes, including TP53 and RBM10. Tumors from Asian never smokers showed a short clonal evolution and presented with heterogeneous repetitive patterns for the inferred mutational order. Importantly, we found that the mutational signature ID2 is a marker of a previously unrecognized mechanism for LUAD evolution. Tumors with ID2 showed short latency and high L1 retrotransposon activity linked to L1 promoter demethylation. These tumors exhibited an aggressive phenotype, characterized by increased genomic instability, elevated hypoxia scores, low burden of neoantigens, propensity to develop metastasis, and poor overall survival. Reactivated L1 retrotransposition-induced mutagenesis can contribute to the origin of the mutational signature ID2, including through the regulation of the transcriptional factor ZNF695, a member of the KZFP family. The complex nature of LUAD evolution creates both challenges and opportunities for screening and treatment plans.
Conflict of interest statement
LBA is a co-founder, CSO, scientific advisory member, and consultant for io9, has equity and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. LBA is also a compensated member of the scientific advisory board of Inocras. LBA’s spouse is an employee of Biotheranostics. LBA declares U.S. provisional applications filed with UCSD with serial numbers: 63/269,033, 63/366,392; 63/289,601; 63/483,237; 63/412,835; and 63/492,348. LBA is also an inventor of a US Patent 10,776,718 for source identification by non-negative matrix factorization. SRY has received consulting fees from AstraZeneca, Sanofi, Amgen, AbbVie, and Sanofi; received speaking fees from AstraZeneca, Medscape, PRIME Education, and Medical Learning Institute. All other authors declare that they have no competing interests.
Figures




References
-
- Cancer facts & figures 2023. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts....
-
- Yang P. et al. Adenocarcinoma of the Lung Is Strongly Associated with Cigarette Smoking: Further Evidence from a Prospective Study of Women. Am. J. Epidemiol. 156, 1114–1122 (2002). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
