This is a preprint.
LANA-Dependent Transcription-Replication Conflicts and R-Loops at the Terminal Repeats (TR) Correlate with KSHV Episome Maintenance
- PMID: 40161765
- PMCID: PMC11952399
- DOI: 10.1101/2025.03.10.642343
LANA-Dependent Transcription-Replication Conflicts and R-Loops at the Terminal Repeats (TR) Correlate with KSHV Episome Maintenance
Update in
-
LANA-dependent transcription-replication conflicts and R-loops at the terminal repeats (TR) correlate with KSHV episome maintenance.PLoS Pathog. 2025 Aug 18;21(8):e1013029. doi: 10.1371/journal.ppat.1013029. eCollection 2025 Aug. PLoS Pathog. 2025. PMID: 40825050 Free PMC article.
Abstract
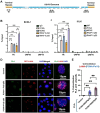
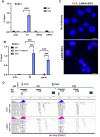
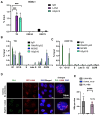
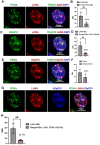
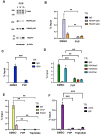
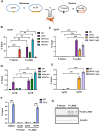
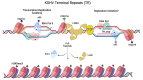
Transcription-replication conflicts frequently occur at repetitive DNA elements involved in genome maintenance functions. The KSHV terminal repeats (TR) function as the viral episome maintenance element when bound by the viral encoded nuclear antigen LANA. Here, we show that transcription-replication conflicts occur at or near LANA binding sites in the TR. We show by proximity ligation assay (PLA) that PCNA and RNAPII colocalize with LANA-nuclear bodies (LANA-NBs). Using DNA-RNA-IP (DRIP) assays with S9.6 antibody, we demonstrate that R-loops form at the TR. We find that these R-loops are also associated with histone H3pS10 a marker for R-loops associated with transcription-replication conflicts. Inhibitors of RNA polymerase eliminated LANA binding to the TR, along with the loss of R-loops and activation associated histone modifications, and the accumulation of heterochromatic marks. We show that LANA can induce all of these features on a plasmid containing 8, but not 2 copies of the TR, correlating strongly with episome maintenance function. Taken together, our study indicates that LANA induces histone modifications associated with RNA and DNA polymerase activity and the formation of R-loops that correlate with episome maintenance function. These findings provide new insights into mechanisms of KSHV episome maintenance during latency and more generally for genome maintenance of repetitive DNA.
Keywords: DNA virus; H3pS10; KSHV; LANA; R-loops; episome maintenance.
Conflict of interest statement
Competing interests: PML declares a competing interest relating to advisory and founding role for Vironika, LLC. All other authors declare they have no competing interests.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
