This is a preprint.
Calcium signals shape metabolic control of H3K27ac and H3K18la to regulate EGA
- PMID: 40161793
- PMCID: PMC11952514
- DOI: 10.1101/2025.03.14.643362
Calcium signals shape metabolic control of H3K27ac and H3K18la to regulate EGA
Abstract
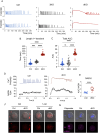
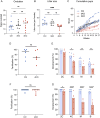
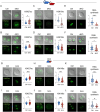
The use of assisted reproductive technologies (ART) has enabled the birth of over 9 million babies; but it is associated with increased risks of negative metabolic outcomes in offspring. Yet, the underlying mechanism remains unknown. Calcium (Ca2+) signals, which initiate embryo development at fertilization, are frequently disrupted in human ART. In mice, abnormal Ca2+ signals at fertilization impair embryo development and adult offspring metabolism. Changes in intracellular Ca2+ drive mitochondrial activity and production of metabolites used by the epigenetic machinery. For example, acetyl-CoA (derived mainly from pyruvate) and lactyl-CoA (derived from lactate) are used for writing H3K27ac and H3K18la marks that orchestrate initiation of development. Using both a genetic mouse model and treatment with ionomycin to raise intracellular Ca2+ of wild-type fertilized eggs, we found that excess Ca2+ at fertilization changes metabolic substrate availability, causing epigenetic changes that impact embryo development and offspring health. Specifically, increased Ca2+ exposure at fertilization led to increased H3K27ac levels and decreased H3K18la levels at the 1-cell (1C) stage, that persisted until the 2-cell (2C) stage. Ultralow input CUT&Tag revealed significant differences in H3K27ac and H3K18la genomic profiles between control and ionomycin groups. In addition, increased Ca2+ exposure resulted in a marked reduction in global transcription at the 1C stage that persisted through the 2C stage due to diminished activity of RNA polymerase I. Excess Ca2+ following fertilization increased pyruvate dehydrogenase activity (enzyme that converts pyruvate to acetyl-CoA) and decreased total lactate levels. Provision of exogenous lactyl-CoA before ionomycin treatment restored H3K18la levels at the 1C and 2C stages and rescued global transcription to control levels. Our findings demonstrate conclusively that Ca2+ dynamics drive metabolic regulation of epigenetic reprogramming at fertilization and alter EGA.
Keywords: DOHaD; Metabolofertility; artificial oocyte activation (AOA); lactylation; zygotic genome activation (ZGA).
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
