This is a preprint.
Muscle-specific increased expression of JAG1 improves skeletal muscle phenotype in dystrophin-deficient mice
- PMID: 40161820
- PMCID: PMC11952387
- DOI: 10.1101/2025.03.12.642857
Muscle-specific increased expression of JAG1 improves skeletal muscle phenotype in dystrophin-deficient mice
Update in
-
Muscle-specific increased expression of JAG1 improves the skeletal muscle phenotype in dystrophin-deficient mice.Proc Natl Acad Sci U S A. 2025 Sep 30;122(39):e2506437122. doi: 10.1073/pnas.2506437122. Epub 2025 Sep 23. Proc Natl Acad Sci U S A. 2025. PMID: 40986346
Abstract
Therapeutic strategies for Duchenne Muscular Dystrophy (DMD) will likely require complementary approaches. One possibility is to explore genetic modifiers that improve muscle regeneration and function. The beneficial effects of the overexpression of Jagged-1 were described in escaper golden retriever muscular dystrophy (GRMD) dogs that had a near-normal life and validated in dystrophin-deficient zebrafish (1). To clarify the underlying biology of JAG1 overexpression in dystrophic muscles, we generated a transgenic mouse (mdx5cv-JAG1) model that lacks dystrophin and overexpresses human JAG1 in striated muscles. Skeletal muscles from mdx5cv-JAG1 and mdx5cv mice were studied at one, four, and twelve-month time points. JAG1 expression in mdx5cv-JAG1 increased by three to five times compared to mdx5cv. Consequently, mdx5cv-JAG1 muscles were significantly bigger and stronger than dystrophic controls, along with an increased number of myofibers. Proteomics data show increased dysferlin in mdx5cv-JAG1 muscles and an association of Nsd1 with the phenotype. Our data supports the positive effect of JAG1 overexpression in dystrophic muscles.
Keywords: Biological Sciences; Duchenne Muscular Dystrophy; Jagged-1; genetic modifier; mouse model; muscles; physiology.
Figures
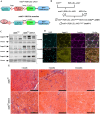
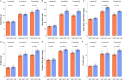

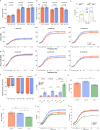
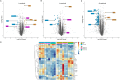
References
-
- Koenig M., et al. , Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50, 509–517 (1987). - PubMed
-
- Hoffman E. P., Brown R. H. Jr., Kunkel L. M., Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987). - PubMed
-
- Emery A. E., Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul Disord 1, 19–29 (1991). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
