This is a preprint.
Ontogeny of the spinal cord dorsal horn
- PMID: 40161840
- PMCID: PMC11952496
- DOI: 10.1101/2025.03.14.643370
Ontogeny of the spinal cord dorsal horn
Abstract
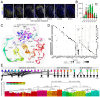
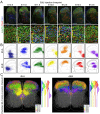
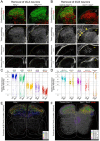
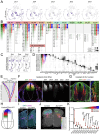
The dorsal horn of the mammalian spinal cord is an exquisite example of form serving function. It is comprised of diverse neuronal populations stacked into laminae, each of which receives different circuit connections and plays specialized roles in behavior. An outstanding question is how this organization emerges during development from an apparently homogeneous pool of neural progenitors. Here, we found that dorsal neurons are diversified by time, with families of related cell types born as temporal cohorts, and by a spatial-molecular gradient that specifies the full array of individual cell types. Excitatory dorsal neurons then settle in a chronotopic arrangement that transforms their progressive birthdates into anatomical order. This establishes the dorsal horn laminae, as these neurons are also required for spatial organization of inhibitory neurons and sensory axons. This work reveals essential ontogenetic principles that shape dorsal progenitors into the diverse cell types and architecture that subserve sensorimotor behavior.
Keywords: Spinal cord; Zic; cell types; development; dorsal horn; interneurons; lamina; ontogeny; structure; temporal specification.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Arokiaraj C.M., Leone M.J., Kleyman M., Chamessian A., Noh M.-C., Phan B.N., Lopes B.C., Corrigan K.A., Cherupally V.K., Yeramosu D., et al. (2024). Spatial, transcriptomic, and epigenomic analyses link dorsal horn neurons to chronic pain genetic predisposition. Cell Rep 43, 114876. 10.1016/j.celrep.2024.114876. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
