This is a preprint.
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli
- PMID: 40161845
- PMCID: PMC11952301
- DOI: 10.1101/2024.10.08.617237
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli
Update in
-
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli.Elife. 2025 Jun 24;14:RP104276. doi: 10.7554/eLife.104276. Elife. 2025. PMID: 40552714 Free PMC article.
Abstract
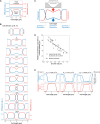
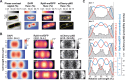
Chromosome segregation is essential for cellular proliferation. Unlike eukaryotes, bacteria lack cytoskeleton-based machinery to segregate their chromosomal DNA (nucleoid). The bacterial ParABS system segregates the duplicated chromosomal regions near the origin of replication. However, this function does not explain how bacterial cells partition the rest (bulk) of the chromosomal material. Furthermore, some bacteria, including Escherichia coli, lack a ParABS system. Yet, E. coli faithfully segregates nucleoids across various growth rates. Here, we provide theoretical and experimental evidence that polysome production during chromosomal gene expression helps compact, split, segregate, and position nucleoids in E. coli through out-of-equilibrium dynamics and polysome exclusion from the DNA meshwork, inherently coupling these processes to biomass growth across nutritional conditions. Halting chromosomal gene expression and thus polysome production immediately stops sister nucleoid migration while ensuing polysome depletion gradually reverses nucleoid segregation. Redirecting gene expression away from the chromosome and toward plasmids causes ectopic polysome accumulations that are sufficient to drive aberrant nucleoid dynamics. Cell width enlargement suggest that the proximity of the DNA to the membrane along the radial axis is important to limit the exchange of polysomes across DNA-free regions, ensuring nucleoid segregation along the cell length. Our findings suggest a self-organizing mechanism for coupling nucleoid segregation to cell growth.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures






References
-
- Bakshi S., Choi H., Mondal J., Weisshaar J.C., 2014. Time-dependent effects of transcription-and translation-halting drugs on the spatial distributions of the E scherichia coli chromosome and ribosomes: Time-dependent drug effects on E. coli chromosome spatial distribution. Mol. Microbiol. 94, 871–887. 10.1111/mmi.12805 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
