Evaluating cell type deconvolution in FFPE breast tissue: application to benign breast disease
- PMID: 40162103
- PMCID: PMC11952925
- DOI: 10.1093/nargab/lqae098
Evaluating cell type deconvolution in FFPE breast tissue: application to benign breast disease
Abstract
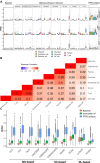
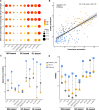
Transcriptome profiling using RNA sequencing (RNA-seq) of bulk formalin-fixed paraffin-embedded (FFPE) tissue blocks is a standard method in biomedical research. However, when used on tissues with diverse cell type compositions, it yields averaged gene expression profiles, complicating biomarker identification due to variations in cell proportions. To address the need for optimized strategies for defining individual cell type compositions from bulk FFPE samples, we constructed single-cell RNA-seq reference data for breast tissue and tested cell type deconvolution methods. Initial simulation experiments showed similar performances across multiple commonly used deconvolution methods. However, the introduction of FFPE artifacts significantly impacted their performances, with a root mean squared error (RMSE) ranging between 0.04 and 0.17. Scaden, a deep learning-based method, consistently outperformed the others, demonstrating robustness against FFPE artifacts. Testing these methods on our 62-sample RNA-seq benign breast disease cohort in which cell type composition was estimated using digital pathology approaches, we found that pre-filtering of the reference data enhanced the accuracy of most methods, realizing up to a 32% reduction in RMSE. To support further research efforts in this domain, we introduce SCdeconR, an R package designed for streamlined cell type deconvolution assessments and downstream analyses.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


References
-
- Hartmann L.C., Sellers T.A., Frost M.H., Lingle W.L., Degnim A.C., Ghosh K., Vierkant R.A., Maloney S.D., Pankratz V.S., Hillman D.W. et al. . Benign breast disease and the risk of breast cancer. N. Engl. J. Med. 2005; 353:229–237. - PubMed
-
- Ludyga N., Grunwald B., Azimzadeh O., Englert S., Hofler H., Tapio S., Aubele M. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch. 2012; 460:131–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

