Inhibition of Ferroptosis Delays Aging and Extends Healthspan Across Multiple Species
- PMID: 40162524
- PMCID: PMC12165029
- DOI: 10.1002/advs.202416559
Inhibition of Ferroptosis Delays Aging and Extends Healthspan Across Multiple Species
Abstract
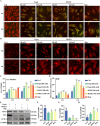
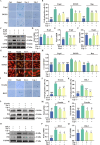
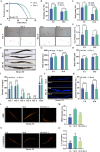
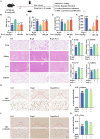
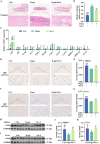
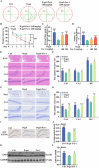
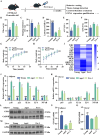
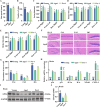
Ferroptosis, a form of iron-dependent cell death, plays a pivotal role in age-related diseases; yet, its impact on cellular senescence and healthspan in mammals remains largely unexplored. This study identifies ferroptosis as a key regulator of cellular senescence, showing that its inhibition can significantly delay aging and extend healthspan across multiple species. During cellular senescence, ferroptosis is progressively exacerbated, marked by increased lipid peroxidation, oxidative stress, and diminished glutathione peroxidase 4 (GPX4) levels. Ferroptosis inducers such as Erastin and RSL3 accelerate senescence; while, inhibitors such as liproxstatin-1 (Lip-1) and ferrostatin-1 (Fer-1) effectively mitigate both chemically and replicatively induced senescence. In vivo, Fer-1 extends lifespan and healthspan in Caenorhabditis elegans, enhances motor function, preserves tissue integrity, and mitigates cognitive decline in both prematurely and naturally aged mice. These effects are attributed to Fer-1's upregulation of GPX4 and inhibition of ferroptosis. Notably, long-term Fer-1 treatment (over 6 months) does not adversely affect body weight or induce aging-related tissue damage but rejuvenates hematological parameters. These findings establish ferroptosis as a critical player in aging dynamics and highlight its inhibition as a promising strategy to extend healthspan and lifespan, providing valuable insights for translational approaches to combat aging and age-related decline.
Keywords: cellular senescence; ferroptosis; ferrostatin‐1; glutathione peroxidase 4; healthspan.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
