Klebsiella pneumoniae evolution in the gut leads to spontaneous capsule loss and decreased virulence potential
- PMID: 40162782
- PMCID: PMC12077207
- DOI: 10.1128/mbio.02362-24
Klebsiella pneumoniae evolution in the gut leads to spontaneous capsule loss and decreased virulence potential
Abstract
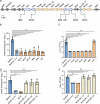
Klebsiella pneumoniae (Kp) is an opportunistic pathogen that poses a major threat in healthcare settings. The gut is a primary Kp reservoir in hospitalized patients, and colonization is a major risk factor for Kp infection. The stability of virulence determinants such as capsule and lipopolysaccharide during gut colonization is largely unexplored. In a murine gut colonization model, we demonstrated that spontaneous capsule loss occurs rapidly but varies by Kp pathotype. A classical Kp strain and a carbapenem-resistant strain of the epidemic sequence type 258 lineage had significant levels (median of 25% and 9.5%, respectively) of capsule loss. In contrast, a hypervirulent strain did not lose capsule to a significant degree (median 0.1%), despite readily losing capsule during in vitro passage. Insertion sequences (ISs) or mutations were found disrupting capsule operon genes of all isolates and in O-antigen encoding genes in a subset of isolates. Mouse-derived acapsular isolates from two pathotypes had significant fitness defects in a murine pneumonia model. Removal of the IS in the capsule operon in a mouse-derived acapsular classical isolate restored capsule production to wild-type levels. Genomic analysis of Klebsiella rectal isolates from hospitalized patients found that 18 of 245 strains (7%) had at least one IS disrupting the capsule operon. Combined, these data indicate that Kp capsule loss can occur during gut colonization in a strain-dependent manner, not only impacting strain virulence but also potentially altering patient infection risk.
Importance: In hospitalized patients, gut colonization by the bacterial pathogen Klebsiella pneumoniae (Kp) is a major risk factor for the development of infections. The genome of Kp varies across isolates, and the presence of certain virulence genes is associated with the ability to progress from colonization to infection. Here, we identified that virulence genes encoding capsule and lipopolysaccharide, which normally protect bacteria from the immune system, are disrupted by mutations during murine gut colonization. These mutations occurred frequently in some isolates of Kp but not others, and these virulence gene mutants from the gut were defective in causing infections. An analysis of 245 human gut isolates demonstrated that this capsule loss also occurred in patients. This work highlights that mutations that decrease virulence occur during gut colonization, the propensity for these mutations differs by isolate, and that stability of virulence genes is important to consider when assessing infection risk in patients.
Keywords: Klebsiella; capsule; evolution; intestinal colonization; lipopolysaccharide; mobile genetic elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA, International Klebsiella Study Group . 2003. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob Agents Chemother 47:3554–3560. doi:10.1128/AAC.47.11.3554-3560.2003 - DOI - PMC - PubMed
-
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi:10.1056/NEJMoa1306801 - DOI - PMC - PubMed
-
- Tsay R-W, Siu LK, Fung C-P, Chang F-Y. 2002. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med 162:1021–1027. doi:10.1001/archinte.162.9.1021 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R01AI125307/National Institute of Allergy and Infectious Diseases
- T32 HG000040/HG/NHGRI NIH HHS/United States
- K99AI175481/National Institute of Allergy and Infectious Diseases
- 1F31AI186288/National Institute of Allergy and Infectious Diseases
- 5T32HG000040/HG/NHGRI NIH HHS/United States
- 32AI007528/National Institute of Allergy and Infectious Diseases
- K99 AI175481/AI/NIAID NIH HHS/United States
- T32 AI007528/AI/NIAID NIH HHS/United States
- R0AI148259/National Institute of Allergy and Infectious Diseases
- R01 AI125307/AI/NIAID NIH HHS/United States
- F31 AI186288/AI/NIAID NIH HHS/United States
- R01 AI148259/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
