Enhancer Hijacking Discovery in Acute Myeloid Leukemia by Pyjacker Identifies MNX1 Activation via Deletion 7q
- PMID: 40162972
- PMCID: PMC12209774
- DOI: 10.1158/2643-3230.BCD-24-0278
Enhancer Hijacking Discovery in Acute Myeloid Leukemia by Pyjacker Identifies MNX1 Activation via Deletion 7q
Abstract
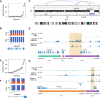
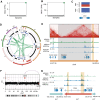
Acute myeloid leukemia (AML) with complex karyotype is characterized by high genomic complexity, including frequent TP53 mutations and chromothripsis. Genomic rearrangements can reposition active enhancers near proto-oncogenes, leading to their aberrant expression; however, a comprehensive understanding of these events in AML is still incomplete. To facilitate the discovery of such "enhancer hijacking" events, we developed Pyjacker, a computational tool, and applied it to 39 AML samples with complex karyotype. Pyjacker identified several enhancer hijacking events in AML patient samples, including aberrant expression of MNX1, which can result from del(7)(q22q36) and is associated with hijacking of a CDK6 enhancer. MNX1 activation occurred in 1.4% of patients with AML and showed significant co-occurrence with BCOR mutations. Through a xenograft mouse model, we demonstrated that MNX1 is required for leukemia cell fitness. Pyjacker is an easy-to-use, accurate, and broadly applicable tool for identifying consequences of genomic events driving tumorigenesis, especially when germline genomic data are missing.
Significance: This study examines the consequences of structural alterations in AML and demonstrates that proto-oncogene activation by enhancer hijacking is an understudied pathomechanism. MNX1 overexpression demonstrates that deletions on chromosome 7q can not only lead to haploinsufficiency but also to activation of oncogenes by enhancer hijacking.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
E. Jahn reports personal fees and other support from AstraZeneca outside the submitted work. F. Brown-Burke reports grants from DKFZ during the conduct of the study. M. Schönung reports other support from Joachim Herz Foundation during the conduct of the study. S.S. Mughal reports grants from DFG (FOR2674) outside the submitted work. B. Brors reports grants from DFG (German Research Council) during the conduct of the study and grants from Hector Foundation II, German Federal Ministry of Research and Education (BMBF): DEEP-HCC/Li-Sym Cancer, and Fritz Thyssen Foundation outside the submitted work. A.-K. Eisfeld reports grants from the NCI and American Cancer Society during the conduct of the study, as well as other support from Karyopharm, personal fees from VJ HemeOnc and Dava Oncology, and Syndax outside the submitted work. L. Bullinger reports grants from Bayer and Jazz Pharmaceuticals and personal fees from Abbvie, Amgen, Astellas, Bristol Myers Squibb, Daiichi Sankyo, Gilead, Glaxo Smith Kline, Janssen, Jazz Pharmaceuticals, Novartis, Otsuka, Pfizer, Roche, Sanofi, and Servier outside the submitted work. K. Döhner reports grants from Astex Pharmaceuticals during the conduct of the study. H. Döhner reports grants from Astex during the conduct of the study. D.B. Lipka reports grants from the German Research Foundation (DFG) during the conduct of the study and personal fees from Infectopharm GmbH outside the submitted work. No disclosures were reported by the other authors.
Figures





References
-
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell 2013;152:1226–36. - PubMed
MeSH terms
Substances
Grants and funding
- R01CA283574-01/American Cancer Society (ACS)
- R01 CA284595/CA/NCI NIH HHS/United States
- SPP1463/Deutsche Forschungsgemeinschaft (DFG)
- HE6233/9-1/Deutsche Forschungsgemeinschaft (DFG)
- HIPO-030/National Center for Tumor Diseases (NCT)
- HE6233/4-2/Deutsche Forschungsgemeinschaft (DFG)
- R01CA284595-01/American Cancer Society (ACS)
- HE6233/10-1/Deutsche Forschungsgemeinschaft (DFG)
- R01 CA283574/CA/NCI NIH HHS/United States
- FOR 2674/Deutsche Forschungsgemeinschaft (DFG)
- SFB 1074/Deutsche Forschungsgemeinschaft (DFG)
- R01 CA262496/CA/NCI NIH HHS/United States
- R01CA262496/American Cancer Society (ACS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

