Genome-wide CRISPR/Cas9 screen reveals factors that influence the susceptibility of tumor cells to NK cell-mediated killing
- PMID: 40164474
- PMCID: PMC11962812
- DOI: 10.1136/jitc-2024-010699
Genome-wide CRISPR/Cas9 screen reveals factors that influence the susceptibility of tumor cells to NK cell-mediated killing
Abstract
Background: Natural killer (NK) cells exhibit potent cytotoxic activity against various cancer cell types. Over the past five decades, numerous methodologies have been employed to elucidate the intricate molecular mechanisms underlying NK cell-mediated tumor control. While significant progress has been made in elucidating the interactions between NK cells and tumor cells, the regulatory factors governing NK cell-mediated tumor cell destruction are not yet fully understood. This includes the diverse array of tumor ligands recognized by NK cells and the mechanisms that NK cells employ to eliminate tumor cells.
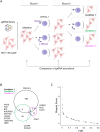
Methods: In this study, we employed a genome-wide CRISPR/Cas9 screening approach in conjunction with functional cytotoxicity assays to delineate the pathways modulating the susceptibility of colon adenocarcinoma HCT-116 cells to NK cell-mediated cytotoxicity.
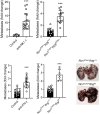
Results: Analysis of guide RNA distribution in HCT-116 cells that survived co-incubation with NK cells identified ICAM-1 as a pivotal player in the NKp44-mediated immune synapse, with NKp44 serving as an activating receptor crucial for the elimination of HCT-116 tumor cells by NK cells. Furthermore, disruption of genes involved in the apoptosis or interferon (IFN)-γ signaling pathways conferred resistance to NK cell attack. We further dissected that NK cell-derived IFN-γ promotes mitochondrial apoptosis in vitro and exerts control over B16-F10 lung metastases in vivo.
Conclusion: Monitoring ICAM-1 levels on the surface of tumor cells or modulating its expression should be considered in the context of NK cell-based therapy. Furthermore, promoting FasL expression on the NK cell surface is reaffirmed as an important strategy to enhance NK cell-mediated tumor killing, offering an additional avenue for therapeutic optimization. Additionally, considering the diffusion properties of IFN-γ, our findings highlight the potential of leveraging NK cell-derived IFN-γ to enhance direct tumor cell killing and facilitate bystander effects via cytokine diffusion, warranting further investigation.
Keywords: Natural killer - NK.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: EV, AF, JG, LG and BR are employees of Innate Pharma. The other authors declare no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
