Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies
- PMID: 40164595
- PMCID: PMC11958643
- DOI: 10.1038/s41467-025-58371-8
Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies
Abstract
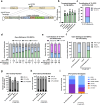
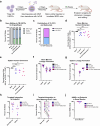
Allogeneic transplantation of CCR5 null hematopoietic stem and progenitor cells (HSPCs) is the only known cure for HIV-1 infection. However, this treatment is limited because of the rarity of CCR5-null matched donors, the morbidities associated with allogeneic transplantation, and the prevalence of HIV-1 strains resistant to CCR5 knockout (KO) alone. Here, we propose a one-time therapy through autologous transplantation of HSPCs genetically engineered ex vivo to produce both CCR5 KO cells and long-term secretion of potent HIV-1 inhibiting antibodies from B cell progeny. CRISPR-Cas9-engineered HSPCs engraft and reconstitute multiple hematopoietic lineages in vivo and can be engineered to express multiple antibodies simultaneously (in pre-clinical models). Human B cells engineered to express each antibody secrete neutralizing concentrations capable of inhibiting HIV-1 pseudovirus infection in vitro. This work lays the foundation for a potential one-time functional cure for HIV-1 through combining the long-term delivery of therapeutic antibodies against HIV-1 and the known efficacy of CCR5 KO HSPC transplantation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.H.P. serves on the scientific advisory board of Allogene Tx and is an advisor to Versant Ventures. M.H.P. has equity in CRISPR Tx and has equity and is a founder of Kamau Therapeutics. M.H.P., W.N.F., and S.E.L. are inventors on a provisional patent application submitted by The Board of Trustees of the Leland Stanford Junior University to the United States Patent and Trademark Office pertaining to the genetic engineering of cells for section of therapeutic antibodies (application number 63/561,249). The remaining authors declare no competing interests.
Figures



References
-
- Joint United Nations Programme on HIV/AIDS. The path that ends AIDS: UNAIDS Global AIDS Update 2023. UNAIDS repor (Joint United Nations Programme on HIV/AIDS, 2023).
-
- Teeraananchai, S., Kerr, S., Amin, J., Ruxrungtham, K. & Law, M. Life expectancy of HIV-positive people after starting combination antiretroviral therapy: a meta-analysis. HIV Med.18, 256–266 (2017). - PubMed
-
- Gandhi, R. T. et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults. JAMA329, 63 (2023). - PubMed
-
- Paterson, D. L. et al. Adherence to protease inhibitor therapy and outcomes in patients with HIV infection. Ann. Intern. Med.133, 21–30 (2000). - PubMed
-
- Barbara, T. Adherence to antiretroviral therapy by human immunodeficiency virus–infected patients. J. Infect. Dis.185, S143–S151 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

