The complexity of tobacco smoke-induced mutagenesis in head and neck cancer
- PMID: 40164736
- PMCID: PMC11985354
- DOI: 10.1038/s41588-025-02134-0
The complexity of tobacco smoke-induced mutagenesis in head and neck cancer
Abstract
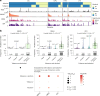
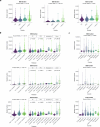
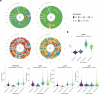
Tobacco smoke, alone or combined with alcohol, is the predominant cause of head and neck cancer (HNC). We explore how tobacco exposure contributes to cancer development by mutational signature analysis of 265 whole-genome sequenced HNC samples from eight countries. Six tobacco-associated mutational signatures were detected, including some not previously reported. Differences in HNC incidence between countries corresponded with differences in mutation burdens of tobacco-associated signatures, consistent with the dominant role of tobacco in HNC causation. Differences were found in the burden of tobacco-associated signatures between anatomical subsites, suggesting that tissue-specific factors modulate mutagenesis. We identified an association between tobacco smoking and alcohol-related signatures, indicating a combined effect of these exposures. Tobacco smoking was associated with differences in the mutational spectra, repertoire of driver mutations in cancer genes and patterns of copy number change. Our results demonstrate the multiple pathways by which tobacco smoke can influence the evolution of cancer cell clones.
© 2025. World Health Organization [2025].
Conflict of interest statement
Competing interests: L.B.A. is a cofounder, chief scientific officer, scientific advisory member and consultant for io9, has equity and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. L.B.A. is also a compensated member of the scientific advisory board of Inocras. L.B.A.’s spouse is an employee of Biotheranostics. E.N.B. and L.B.A. declare a US provisional patent application filed with UCSD with serial numbers 63/289,601, 63/269,033 and 63/483,237. A.A. and L.B.A. declare a US provisional patent application filed with UCSD with serial number 63/366,392. L.B.A. also declares US provisional application 63/412,835 and international application PCT/US2023/010679 filed with UCSD and is also an inventor of a US Patent 10,776,718 for source identification by non-negative matrix factorization. All other authors declare no competing interests.
Figures














Update of
-
The Complexity of Tobacco Smoke-Induced Mutagenesis in Head and Neck Cancer.medRxiv [Preprint]. 2024 Apr 17:2024.04.15.24305006. doi: 10.1101/2024.04.15.24305006. medRxiv. 2024. Update in: Nat Genet. 2025 Apr;57(4):884-896. doi: 10.1038/s41588-025-02134-0. PMID: 38699364 Free PMC article. Updated. Preprint.
References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Simard, E. P., Torre, L. A. & Jemal, A. International trends in head and neck cancer incidence rates: differences by country, sex and anatomic site. Oral Oncol.50, 387–403 (2014). - PubMed
MeSH terms
Grants and funding
- R01CA269919-01/U.S. Department of Health and Human Services (U.S. Department of Health & Human Services)
- C98/A24032/Cancer Research UK (CRUK)
- R01 ES032547/ES/NIEHS NIH HHS/United States
- U01 CA290479/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA269919/CA/NCI NIH HHS/United States
- 825771/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 001/WHO_/World Health Organization/International
- 1U01CA290479-01/U.S. Department of Health and Human Services (U.S. Department of Health & Human Services)
- R01ES032547-01/U.S. Department of Health and Human Services (U.S. Department of Health & Human Services)

