Liver TET1 promotes metabolic dysfunction-associated steatotic liver disease
- PMID: 40164757
- PMCID: PMC12081649
- DOI: 10.1038/s44321-025-00224-4
Liver TET1 promotes metabolic dysfunction-associated steatotic liver disease
Abstract
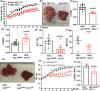
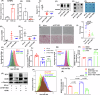
Global hepatic DNA methylation change has been linked to human patients with metabolic dysfunction-associated steatotic liver disease (MASLD). DNA demethylation is regulated by the TET family proteins, whose enzymatic activities require 2-oxoglutarate (2-OG) and iron that both are elevated in human MASLD patients. We aimed to investigate liver TET1 in MASLD progression. Depleting TET1 using two different strategies substantially alleviated MASLD progression. Knockout (KO) of TET1 slightly improved diet induced obesity and glucose homeostasis. Intriguingly, hepatic cholesterols, triglycerides, and CD36 were significantly decreased upon TET1 depletion. Consistently, liver specific TET1 KO led to improvement of MASLD progression. Mechanistically, TET1 promoted CD36 expression through transcriptional upregulation via DNA demethylation control. Overexpression of CD36 reversed the impacts of TET1 downregulation on fatty acid uptake in hepatocytes. More importantly, targeting TET1 with a small molecule inhibitor significantly suppressed MASLD progression. Conclusively, liver TET1 plays a deleterious role in MASLD, suggesting the potential of targeting TET1 in hepatocytes to suppress MASLD.
Keywords: 5-Hydroxymethylcytosine; Alcoholic Liver Disease; Epigenetics; Fatty Liver; Nonalcoholic Fatty Liver Disease.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. Dr. Xiao-Ming Yin is the Board of Advisor, NanoPin Technologies, New Orleans, LA. The NanoPin Technologies has no role in this study. All other authors have no competing interests.
Figures





References
-
- Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A (2012) The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol 56:952–964 - PubMed
MeSH terms
Substances
Grants and funding
- DK135664/HHS | NIH | NIDDK | Division of Diabetes, Endocrinology, and Metabolic Diseases (DEM)
- R01AA030424/HHS | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- R01AA023190/HHS | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- P20GM109035/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 AA023190/AA/NIAAA NIH HHS/United States
- R01 DK135664/DK/NIDDK NIH HHS/United States
- R01 AA030424/AA/NIAAA NIH HHS/United States
- P20 GM109035/GM/NIGMS NIH HHS/United States
- R21AA028576/HHS | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- 2017 AASLDF Pinnacle Research Development Award/American Association for the Study of Liver Diseases (AASLD)
- R21 AA028576/AA/NIAAA NIH HHS/United States
- R01 AA031497/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

