Parasite and vector circadian clocks mediate efficient malaria transmission
- PMID: 40164831
- PMCID: PMC11964930
- DOI: 10.1038/s41564-025-01949-1
Parasite and vector circadian clocks mediate efficient malaria transmission
Abstract
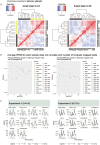
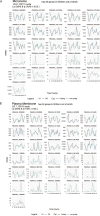
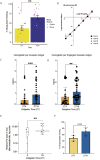
Malaria transmission begins when Anopheles mosquitos deposit saliva and Plasmodium parasites during a bloodmeal. As Anopheles mosquitos are nocturnal, we investigated whether their salivary glands are under circadian control, anticipating bloodmeals and modulating parasite biology for host encounters. Here we show that approximately half of the mosquito salivary gland transcriptome, particularly genes essential for efficient bloodmeals such as anti-blood clotting factors, exhibits circadian expression. Furthermore, measuring haemoglobin levels, we demonstrate that mosquitos prefer to feed and ingest more blood at nighttime. Notably, we show a substantial subset of the salivary-gland-resident parasite transcriptome cycling throughout the day, indicating that this stage is not transcriptionally quiescent. Among the sporozoite genes undergoing rhythmic expression are those involved in parasite motility, potentially modulating the ability to initiate infection at different times of day. Our findings suggest a circadian tripartite relationship between the vector, parasite and mammalian host that together modulates malaria transmission.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- World Malaria Report 2022 (WHO, 2022).
-
- Jones, M. D., Ford, M. G. & Gillett, J. D. Light-on and light-off effects on the circadian flight activity in the mosquito Anopheles gambiae. Nature211, 871–872 (1966). - PubMed
MeSH terms
Grants and funding
- searle scholars/Kinship Foundation
- HR17-00264-PoEMM/"la Caixa" Foundation (Caixa Foundation)
- K99 GM132557/GM/NIGMS NIH HHS/United States
- ALTF-84-2023/European Molecular Biology Organization (EMBO)
- 1K99GM132557/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

