The biochemical mechanism of Rho GTPase membrane binding, activation and retention in activity patterning
- PMID: 40164947
- PMCID: PMC12048676
- DOI: 10.1038/s44318-025-00418-z
The biochemical mechanism of Rho GTPase membrane binding, activation and retention in activity patterning
Abstract
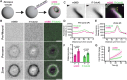
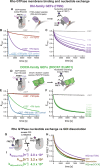
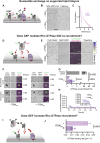
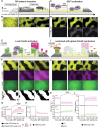
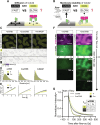
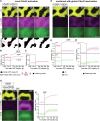
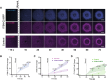
Rho GTPases form plasma membrane-associated patterns that control the cytoskeleton during cell division, morphogenesis, migration, and wound repair. Their patterning involves transitions between inactive cytosolic and active membrane-bound states, regulated by guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs). However, the relationships between these transitions and role of different regulators remain unclear. We developed a novel reconstitution approach to study Rho GTPase patterning with all major GTPase regulators in a biochemically defined system. We show that Rho GTPase dissociation from RhoGDI is rate-limiting for its membrane association. Rho GTPase activation occurs after membrane insertion, which is unaffected by GEF activity. Once activated, Rho GTPases are retained at the membrane through effector interactions, essential for their enrichment at activation sites. Thus, high cytosolic levels of RhoGDI-bound GTPases ensure a constant supply of inactive GTPases for the membrane, where GEF-mediated activation and effector binding stabilize them. These results delineate the route by which Rho GTPase patterns are established and define stage-dependent roles of its regulators.
Keywords: Cell Polarity; Membrane Signaling; Rho GTPases; Self-organization; Small G Proteins.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures







References
-
- Bement WM, Goryachev AB, Miller AL, von Dassow G (2024) Patterning of the cell cortex by Rho GTPases. Nat Rev Mol Cell Biol 25:290–308 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

