An Agrin-YAP/TAZ Rigidity Sensing Module Drives EGFR-Addicted Lung Tumorigenesis
- PMID: 40165020
- PMCID: PMC12120797
- DOI: 10.1002/advs.202413443
An Agrin-YAP/TAZ Rigidity Sensing Module Drives EGFR-Addicted Lung Tumorigenesis
Abstract
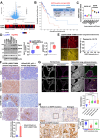
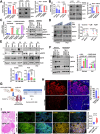
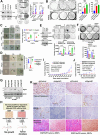
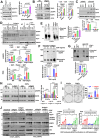
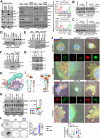
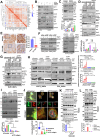
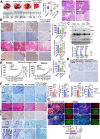
Despite epidermal growth factor receptor (EGFR) is a pivotal oncogene for several cancers, including lung adenocarcinoma (LUAD), how it senses extracellular matrix (ECM) rigidity remain elusive in the context of the increasing role of tissue rigidity on various hallmarks of cancer development. Here it is shown that EGFR dictates tumorigenic agrin expression in lung cancer cell lines, genetically engineered EGFR-driven mouse models, and human specimens. Agrin expression confers substrate stiffness-dependent oncogenic attributes to EGFR-reliant cancer cells. Mechanistically, agrin mechanoactivates EGFR through epidermal growth factor (EGF)-dependent and independent modes, thereby sensitizing its activity toward localized cancer cell-ECM adherence and bulk rigidity by fostering interactions with integrin β1. Notably, a feed-forward loop linking agrin-EGFR rigidity response to YAP-TEAD mechanosensing is essential for tumorigenesis. Together, the combined inhibition of EGFR-YAP/TEAD may offer a strategy to reduce lung tumorigenesis by disrupting agrin-EGFR mechanotransduction, uncovering a therapeutic vulnerability for EGFR-addicted lung cancers.
Keywords: EGFR, extracellular matrix; YAP/TAZ; agrin; hippo pathway; lung cancer.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Gridelli C., Rossi A., Carbone D. P., Guarize J., Karachaliou N., Mok T., Petrella F., Spaggiari L., Rosell R., Nat. Rev. Dis. Primers 2015, 1, 15009. - PubMed
-
- Hirsch F. R., Varella‐Garcia M., P. A. Bunn, Jr. , Di Maria M. V., Veve R., Bremmes R. M., Baron A. E., Zeng C., Franklin W. A., J. Clin. Oncol. 2003, 21, 3798. - PubMed
-
- a) Sharma S. V., Bell D. W., Settleman J., Haber D. A., Nat. Rev. Cancer 2007, 7, 169; - PubMed
- b) Shigematsu H., Lin L., Takahashi T., Nomura M., Suzuki M., Wistuba I. I., Fong K. M., Lee H., Toyooka S., Shimizu N., Fujisawa T., Feng Z., Roth J. A., Herz J., Minna J. D., Gazdar A. F., J. Natl. Cancer Inst. 2005, 97, 339. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
