Integrated bioinformatics and experiment validation reveal cuproptosis-related biomarkers and therapeutic targets in sepsis-induced myocardial dysfunction
- PMID: 40165133
- PMCID: PMC11956201
- DOI: 10.1186/s12879-025-10822-9
Integrated bioinformatics and experiment validation reveal cuproptosis-related biomarkers and therapeutic targets in sepsis-induced myocardial dysfunction
Abstract
Background: Sepsis-induced myocardial dysfunction (SIMD) is a serious sepsis complication with high mortality, yet current diagnostic and therapeutic approaches remain limited. The lack of early, specific biomarkers and effective treatments necessitates exploration of novel mechanisms. Recently, cuproptosis has been implicated in various diseases, but its role in SIMD is unclear. This study aimed to identify cuproptosis-related biomarkers and potential therapeutic agents, supported by animal model validation.
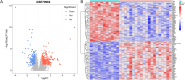
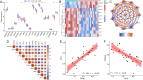
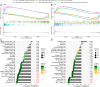
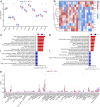
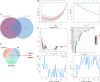
Methods: Four GEO datasets (GSE79962, GSE267388, GSE229925, GSE229298) were analyzed using Limma and WGCNA to identify overlapping genes from differentially expressed genes (DEGs), cuproptosis-related DEGs (DE-CRGs), and module-associated genes. Gene Set Enrichment Analysis (GSEA) and single-sample GSEA (ssGSEA) were performed to assess biological functions and immune cell infiltration, respectively. ceRNA and transcription factor networks were constructed to explore gene regulatory mechanisms, while consensus clustering was employed to define cuproptosis-related subtypes. Diagnostic genes were selected through SVM-RFE, LASSO, and random forest models. Additionally, potential gene-targeting agents were predicted using drug-gene interaction analysis. The findings were validated in SIMD animal models through qPCR and immunohistochemical analysis to confirm gene expression.
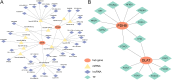
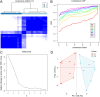
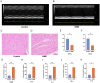
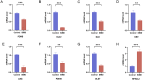
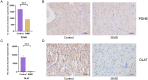
Results: PDHB and DLAT emerged as key cuproptosis-related biomarkers. GSEA indicated upregulation of oxidative phosphorylation and downregulation of chemokine signaling. ssGSEA revealed negative correlations with several immune cell types. A ceRNA network (51 nodes, 56 edges) was constructed. Machine learning identified PDHB, NDUFA9, and TIMMDC1 as diagnostic genes, with PDHB showing high accuracy (AUC = 0.995 in GSE79962; AUC = 0.960, 0.864, and 0.984 in external datasets). Using the DSigDB database, we predicted six drugs that exhibit significant binding activity with PDHB. qPCR and immunohistochemistry confirmed reduced PDHB and DLAT expression in SIMD animal models.
Conclusion: This study identifies PDHB and DLAT as cuproptosis-related biomarkers, addressing the diagnostic and therapeutic gaps in SIMD by unveiling novel molecular insights for early intervention and targeted treatment.
Clinical trial number: Not applicable.
Keywords: Biomarkers; Cuproptosis-related genes; Machine learning; Molecular mechanism; Sepsis-induced myocardial dysfunction; Therapeutic targets.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Studies involving animals have been reviewed and approved by the Ethics Committee of the Affiliated Hospital of Southwest Medical University. NO.:20231016-013. Consent for publication: Not Applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Parrillo JE, Parker MM, Natanson C, Suffredini AF, Danner RL, Cunnion RE, Ognibene FP. Septic shock in humans. Advances in the Understanding of pathogenesis, cardiovascular dysfunction, and therapy. Ann Intern Med. 1990;113(3):227–42. - PubMed
-
- Hollenberg SM, Singer M. Pathophysiology of sepsis-induced cardiomyopathy. Nat Rev Cardiol. 2021;18(6):424–34. - PubMed
MeSH terms
Substances
Grants and funding
- 2020ZRQNA004/scientific project of the Southwest Medical University
- S23007/Science Foundation of The Sichuan Medical Association Project
- 2018099/Doctoral Research Foundation of the Affiliated Hospital of Southwest Medical University
- S2024010/Medical Research of the Sichuan Medical Association Project
LinkOut - more resources
Full Text Sources
Medical

