tesG expression as a potential clinical biomarker for chronic Pseudomonas aeruginosa pulmonary biofilm infections
- PMID: 40165235
- PMCID: PMC11959726
- DOI: 10.1186/s12916-025-04009-x
tesG expression as a potential clinical biomarker for chronic Pseudomonas aeruginosa pulmonary biofilm infections
Abstract
Background: Pseudomonas aeruginosa infections in the lungs affect millions of children and adults worldwide. To our knowledge, no clinically validated prognostic biomarkers for chronic pulmonary P. aeruginosa infections exist. Therefore, this study aims to identify potential prognostic markers for chronic P. aeruginosa biofilm lung infections.
Methods: Here, we screened the expression of 11 P. aeruginosa regulatory genes (tesG, algD, lasR, lasA, lasB, pelB, phzF, rhlA, rsmY, rsmZ, and sagS) to identify associations between clinical status and chronic biofilm infection.
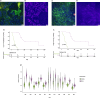
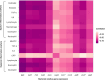
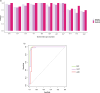
Results: RNA was extracted from 210 sputum samples from patients (n = 70) with chronic P. aeruginosa lung infections (mean age; 29.3-56.2 years; 33 female). Strong biofilm formation was correlated with prolonged hospital stays (212.2 days vs. 44.4 days) and increased mortality (46.2% (18)). Strong biofilm formation is associated with increased tesG expression (P = 0.001), influencing extended intensive care unit (P = 0.002) or hospitalisation stays (P = 0.001), pneumonia risk (P = 0.006), and mortality (P = 0.001). Notably, tesG expression is linked to the modulation of systemic and sputum inflammatory responses and predicts biofilm biomass.
Conclusions: This study provides the first clinical dataset of tesG expression levels as a predictive biomarker for chronic P. aeruginosa pulmonary infections.
Keywords: Pseudomonas aeruginosa; Chronic infections; Chronic respiratory diseases; Clinical biomarker; Pulmonary biofilm; Respiratory infections; TesG expression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study protocol was approved (IRB No. 414/60) by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, and was performed in accordance with the ethical standards as established in the 1964 Declaration of Helsinki and its later amendments and comparable ethical standards. For this retrospective, non-interventional study of pseudonymised clinical isolates, the requirement for informed consent from patients was waived by the Institutional Review Board (IRB No. 414/60) of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. Competing interests: The authors declare no competing interests.
Figures

References
-
- Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol. 2021;19(5):331–42. - PubMed
-
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10(12):841–51. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

