Translation of bi-directional transcripts enhances MHC-I peptide diversity
- PMID: 40165968
- PMCID: PMC11956742
- DOI: 10.3389/fimmu.2025.1554561
Translation of bi-directional transcripts enhances MHC-I peptide diversity
Abstract
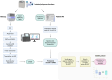
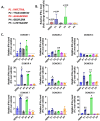
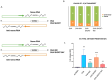
Antisense transcripts play an important role in generating regulatory non-coding RNAs but whether these transcripts are also translated to generate functional peptides remains poorly understood. In this study, RNA sequencing and six-frame database generation were combined with mass spectrometry analysis of peptides isolated from polysomes to identify Nascent Pioneer Translation Products (Na-PTPs) originating from alternative reading frames of bi-directional transcripts. Two Na-PTP originating peptides derived from antisense strands stimulated CD8+ T cell proliferation when presented to peripheral blood mononuclear cells (PBMCs) from nine healthy donors. Importantly, an antigenic peptide derived from the reverse strand of two cDNA constructs was presented on MHC-I molecules and induced CD8+ T cell activation. The results demonstrate that three-frame translation of bi-directional transcripts generates antigenic peptide substrates for the immune system. This discovery holds significance for understanding the origin of self-discriminating peptide substrates for the major histocompatibility class I (MHC-I) pathway and for enhancing immune-based therapies against infected or transformed cells.
Keywords: MHC-I epitope; Pioneer Translation Products; bi-directional transcripts; bi-directional translation; reverse strand antigenic peptides.
Copyright © 2025 Zavadil, Henek, Habault, Chemali, Tovar-Fernandez, Daskalogianni, Malbert-Colas, Wang, Gnanasundram, Vojtesek, Hernychova, Apcher and Fahraeus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

