Exploring chemical space for "druglike" small molecules in the age of AI
- PMID: 40166082
- PMCID: PMC11955463
- DOI: 10.3389/fmolb.2025.1553667
Exploring chemical space for "druglike" small molecules in the age of AI
Abstract
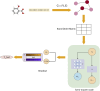
The announcement of 2024 Nobel Prize in Chemistry to Alphafold has reiterated the role of AI in biology and mainly in the domain of "drug discovery". Till few years ago, structure-based drug design (SBDD) has been the preferred experimental design in many academic and pharmaceutical R and D divisions for developing novel therapeutics. However, with the advent of AI, the drug design field especially has seen a paradigm shift in its R&D across platforms. If "drug design" is a game, there are two main players, the small molecule drug and its target biomolecule, and the rules governing the game are mainly based on the interactions between these two players. In this brief review, we will be discussing our efforts in improving the state-of-the-art technology with respect to small molecules as well as in understanding the rules of the game. The review is broadly divided into five sections with the first section introducing the field and the challenges faced and the role of AI in this domain. In the second section, we describe some of the existing small molecule libraries developed in our labs and follow-up this section with a more recent knowledge-based resource available for public use. In section four, we describe some of the screening tools developed in our laboratories and are available for public use. Finally, section five delves into how domain knowledge is improving the utilization of AI in drug design. We provide three case studies from our work to illustrate this work. Finally, we conclude with our thoughts on the future scope of AI in drug design.
Keywords: BIMP; artificial intelligence; computer aided drug design (CADD); machine learning (ML); small molecules.
Copyright © 2025 Kattuparambil, Chaurasia, Shekhar, Srinivasan, Mondal, Aduri and Jayaram.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Appell K., Baldwin J. J., Egan J. W. (2001). “Combinatorial chemistry and high-throughput screening in drug discovery and development,” in Handbook of modern pharmaceutical analysis (Academic Press; ), 24–56.
-
- Bertin P., Rector-Brooks J., Sharma D., Gaudelet T., Anighoro A., Gross T., et al. (2022). RECOVER: sequential model optimization platform for combination drug repurposing identifies novel synergistic compounds in vitro . ArXiv. Available online at: http://arxiv.org/abs/2202.04202.
-
- Bhat B. S., Srinivasan A., Dash T., Krishnan S. R., Vig L., Roy A., et al. (2024). “Generating novel leads for drug discovery using LLMs with logical feedback,” in Proceedings of the AAAI conference on artificial intelligence. USA, February 20–27, 2024, Available online at: https://ojs.aaai.org/index.php/AAAI/article/view/27751.
Publication types
LinkOut - more resources
Full Text Sources

