The comprehensive assessment of epigenetics changes during skin development
- PMID: 40166100
- PMCID: PMC11955039
- DOI: 10.1016/j.fmre.2022.08.021
The comprehensive assessment of epigenetics changes during skin development
Abstract
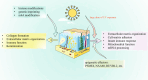
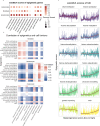
Epigenetic regulation is critical to multiple physiological and pathological processes. However, little is known regarding the epigenetic changes during neonatal skin development and skin aging, and in response to ultraviolet (UV) exposure. The transcriptomes of human skin samples from different ages or irradiated with different types and doses of UV light were analyzed using R (version 4.0.3) software. The epigenetic landscape of the skin, including histone modifications, genetic imprinting and m6A modification, which are mainly involved in collagen formation, extracellular matrix organization, immune function and keratinization, underwent significant changes during neonatal to adult development. Epigenetic effectors such as IGF2BP2, GATA2, GATA3, CPA4 and CDK1 were significantly correlated with extracellular matrix organization, and VEGFA, CDK1 and PRKCB with skin immune function. The m6A readers such as IGF2BP2, IGF2BP3, HNRNPA2B1 and EIF3G showed significant correlation with extracellular matrix organization, metabolism, or antigen processing and presentation. Small doses of UV exposure only induced changes in the expression levels of some epigenetic effectors, without any significant effect on the overall epigenetic landscape. However, the minimal erythema dose of UV exposure altered multiple epigenetic effectors regulating extracellular matrix organization, cell-matrix adhesion, innate immune response, mitochondrial function and mRNA processing. In addition, epigenetic changes following UV exposure were more pronounced in the elderly skin compared to the younger skin. In conclusion, histone modifications, genetic imprinting and m6A modification play critical roles during skin development, and a large dose of UV exposure can significantly change the expression of multiple epigenetic effectors.
Keywords: Aging; Epigenetics; Skin development; Ultraviolet; m6A.
© 2022 The Authors. Publishing Services by Elsevier B.V. on behalf of KeAi Communications Co. Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures






Similar articles
-
Downregulation of carnitine acetyltransferase by promoter hypermethylation regulates ultraviolet-induced matrix metalloproteinase-1 expression in human dermal fibroblasts.J Dermatol Sci. 2024 Nov;116(2):70-77. doi: 10.1016/j.jdermsci.2024.09.005. Epub 2024 Sep 27. J Dermatol Sci. 2024. PMID: 39443271
-
Ultraviolet irradiation-induced inhibition of histone deacetylase 4 increases the expression of matrix metalloproteinase-1 but decreases that of type I procollagen via activating JNK in human dermal fibroblasts.J Dermatol Sci. 2021 Feb;101(2):107-114. doi: 10.1016/j.jdermsci.2020.11.008. Epub 2020 Dec 3. J Dermatol Sci. 2021. PMID: 33309320
-
A dynamic reversible RNA N6 -methyladenosine modification: current status and perspectives.J Cell Physiol. 2019 Jun;234(6):7948-7956. doi: 10.1002/jcp.28014. Epub 2019 Jan 15. J Cell Physiol. 2019. PMID: 30644095 Review.
-
Chronic UVB- and all-trans retinoic-acid-induced qualitative and quantitative changes in hairless mouse skin.J Photochem Photobiol B. 1995 May;28(2):125-35. doi: 10.1016/1011-1344(94)07080-8. J Photochem Photobiol B. 1995. PMID: 7636633
-
Epigenetics and environmental health.Front Med. 2024 Aug;18(4):571-596. doi: 10.1007/s11684-023-1038-2. Epub 2024 May 28. Front Med. 2024. PMID: 38806988 Review.
References
-
- Stricker S.H., Köferle A., Beck S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017;18(1):51–66. - PubMed
-
- Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489–499. - PubMed
-
- Long H., Yin H., Wang L., et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J. Autoimmun. 2016;74:118–138. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

