This is a preprint.
Spatial patterning of fibroblast TGFβ signaling underlies treatment resistance in rheumatoid arthritis
- PMID: 40166167
- PMCID: PMC11956964
- DOI: 10.1101/2025.03.14.642821
Spatial patterning of fibroblast TGFβ signaling underlies treatment resistance in rheumatoid arthritis
Abstract
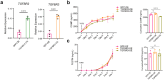
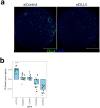
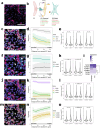
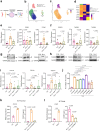
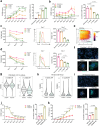
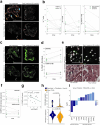
Treatment-refractory rheumatoid arthritis (RA) is a major unmet need, and the mechanisms driving treatment resistance are poorly understood. To identify molecular determinants of RA non-remission, we performed spatial transcriptomic profiling on pre- and post-treatment synovial tissue biopsies from treatment naïve patients who received conventional DMARDs or adalimumab for 6 months. In the baseline biopsies of non-remission patients, we identified significant expansion of fibrogenic fibroblasts marked by high expression of COMP, a fibrosis-associated extracellular matrix protein. COMPhi fibroblasts localized to perivascular niches that, unexpectedly, served as transcriptional hubs for TGFβ activity. We identified endothelial-derived Notch signaling as an upstream regulator of fibroblast TGFβ signaling via its dual role in driving TGFβ isoform expression and suppressing TGFβ receptors, generating a proximal-distal gradient of TGFβ activity. Further, disruption of steady-state Notch signaling in vitro enabled fibrogenic fibroblast activation. Analysis of post-treatment biopsies revealed marked expansion of COMPhi fibroblasts in non-remission RA patients, despite evidence of successful immune cell depletion, suggesting a spatiotemporal process of fibrogenic remodeling linked to treatment resistance. Collectively, our data implicates targeting of TGFβ signaling to prevent exuberant synovial tissue fibrosis as a potential therapeutic strategy for refractory RA.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
