This is a preprint.
GMCL1 Controls 53BP1 Stability and Modulates Paclitaxel Sensitivity in Cancer
- PMID: 40166203
- PMCID: PMC11957010
- DOI: 10.1101/2025.03.18.643855
GMCL1 Controls 53BP1 Stability and Modulates Paclitaxel Sensitivity in Cancer
Abstract
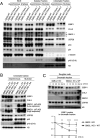
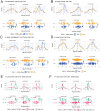
The Mitotic Surveillance Pathway (MSP) monitors the duration of M-phase. Prolonged mitosis, caused by spindle attachment defects or microtubule-targeting drugs such as the taxane paclitaxel, induces the formation of the ternary "mitotic stopwatch" complex consisting of 53BP1, USP28, and p53. This event protects p53 from degradation, resulting in cell cycle arrest or apoptosis in daughter cells. In paclitaxel-resistant cancers, cells bypass the MSP, enabling unchecked proliferation and survival, although the underlying mechanisms remain unknown. Here, we demonstrate that 53BP1 physically interacts with GMCL1 but not its paralog, GMCL2, and we mapped the interaction regions on both proteins. CRL3GMCL1 functions as a ubiquitin ligase that targets 53BP1 for degradation during M phase, impacting p53 levels in daughter cells. High GMCL1 expression significantly correlates with resistance to paclitaxel in cancer cell lines with wild-type p53, including endometrial, breast, and upper aerodigestive tract cancer cells. Loss of GMCL1 restores paclitaxel sensitivity in p53 expressing cells but not in p53 deficient cells. We propose that in cancers with high GMCL1 levels, the CRL3GMCL1-mediated degradation of 53BP1 prevents the formation of the mitotic stopwatch complex, leading to p53 degradation and sustained proliferation. Finally, our results indicate that GMCL1 inhibition represents a novel strategy to restore taxane sensitivity in resistant cancers.
Keywords: 53BP1; GMCL1; mitotic stopwatch; p53; prolonged mitosis; protein degradation; ubiquitin.
Conflict of interest statement
DECLARATION OF INTERESTS MP is or has been an advisor for SEED Therapeutics, CullGen, Deargen, Kymera Therapeutics, Lumanity, Serinus Biosciences, Sibylla Biotech, Triana Biomedicines, and Umbra Therapeutics. He also has financial interests in CullGen, Kymera Therapeutics, SEED Therapeutics, Thermo Fisher Scientific, and Triana Biomedicines. The other authors declare no conflict of interest.
Figures


References
-
- Contadini C., Monteonofrio L., Virdia I., Prodosmo A., Valente D., Chessa L., Musio A., Fava L.L., Rinaldo C., Di Rocco G., and Soddu S. (2019). p53 mitotic centrosome localization preserves centrosome integrity and works as sensor for the mitotic surveillance pathway. Cell Death Dis 10, 850. 10.1038/s41419-019-2076-1. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
