This is a preprint.
In vivo validation of the palmitoylation cycle as a therapeutic target in NRAS-mutant cancer
- PMID: 40166265
- PMCID: PMC11957127
- DOI: 10.1101/2025.03.20.644389
In vivo validation of the palmitoylation cycle as a therapeutic target in NRAS-mutant cancer
Abstract
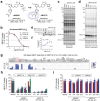
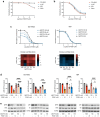
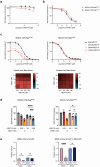
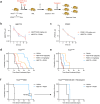
Normal and oncogenic Ras proteins are functionally dependent on one or more lipid modifications1,2. Whereas K-Ras4b farnesylation is sufficient for stable association with the plasma membrane, farnesylated H-Ras, K-Ras4a, and N-Ras traffic to the Golgi where they must undergo palmitoylation before regulated translocation to cell membranes. N-Ras palmitoylation by the DHHC family of palmitoyl acyl transferases (PATs) and depalmitoylation by ABHD17 serine hydrolases is a dynamic process that is essential for the growth of acute myeloid leukemias (AMLs) harboring oncogenic NRAS mutations3-6. Here, we have tested whether co-targeting ABHD17 enzymes and Ras signal output would cooperatively inhibit the proliferation and survival of NRAS-mutant AMLs while sparing normal tissues that retain K-Ras4b function. We show that ABD778, a potent and selective ABHD17 inhibitor with in vivo activity, selectively reduces the growth of NRAS-mutant AML cells in vitro and is synergistic with the allosteric MEK inhibitor PD0325901 (PD901)7,8. Similarly, ABD778 and PD901 significantly extended the survival of recipient mice transplanted with three independent primary mouse AMLs harboring an oncogenic Nras G12D driver mutation. Resistant leukemias that emerged during continuous drug treatment acquired by-pass mutations that confer adaptive drug resistance and increase mitogen activated protein kinase (MAPK) signal output. ABD778 augmented the anti-leukemia activity of the pan-PI3 kinase inhibitor pictilisib9, the K/N-RasG12C inhibitor sotorasib10, and the FLT3 inhibitor gilteritinib11. Co-treatment with ABD778 and gilteritinib restored drug sensitivity in a patient-derived xenograft model of adaptive resistance to FLT3 inhibition. These data validate the palmitoylation cycle as a promising therapeutic target in AML and support exploring it in other NRAS-mutant cancers.
Conflict of interest statement
Competing interests Nhi Ngo, Dylan M. Herbst, Radu M. Suciu, Carlos Guijas, Kimia Sedighi, Taylor Andalis, Elysia Roche, Boer Xie, and Micah J. Niphakis are full-time employees of Lundbeck. The authors declare no other competing interests.
Figures

Similar articles
-
Preclinical efficacy of MEK inhibition in Nras-mutant AML.Blood. 2014 Dec 18;124(26):3947-55. doi: 10.1182/blood-2014-05-574582. Epub 2014 Oct 31. Blood. 2014. PMID: 25361812 Free PMC article.
-
ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth.Nat Chem Biol. 2021 Aug;17(8):856-864. doi: 10.1038/s41589-021-00785-8. Epub 2021 Apr 29. Nat Chem Biol. 2021. PMID: 33927411 Free PMC article.
-
RAB27B controls palmitoylation-dependent NRAS trafficking and signaling in myeloid leukemia.J Clin Invest. 2023 Jun 15;133(12):e165510. doi: 10.1172/JCI165510. J Clin Invest. 2023. PMID: 37317963 Free PMC article.
-
Blockade of mutant RAS oncogenic signaling with a special emphasis on KRAS.Pharmacol Res. 2021 Oct;172:105806. doi: 10.1016/j.phrs.2021.105806. Epub 2021 Aug 24. Pharmacol Res. 2021. PMID: 34450320 Review.
-
A New View of Ras Isoforms in Cancers.Cancer Res. 2016 Jan 1;76(1):18-23. doi: 10.1158/0008-5472.CAN-15-1536. Epub 2015 Dec 10. Cancer Res. 2016. PMID: 26659836 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
