This is a preprint.
Genetic diversity and regulatory features of human-specific NOTCH2NL duplications
- PMID: 40166283
- PMCID: PMC11956922
- DOI: 10.1101/2025.03.14.643395
Genetic diversity and regulatory features of human-specific NOTCH2NL duplications
Abstract
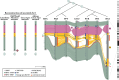
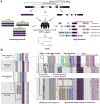
NOTCH2NL (NOTCH2-N-terminus-like) genes arose from incomplete, recent chromosome 1 segmental duplications implicated in human brain cortical expansion. Genetic characterization of these loci and their regulation is complicated by the fact they are embedded in large, nearly identical duplications that predispose to recurrent microdeletion syndromes. Using nearly complete long-read assemblies generated from 67 human and 12 ape haploid genomes, we show independent recurrent duplication among apes with functional copies emerging in humans ~2.1 million years ago. We distinguish NOTCH2NL paralogs present in every human haplotype (NOTCH2NLA) from copy number variable ones. We also characterize large-scale structural variation, including gene conversion, for 28% of haplotypes leading to a previously undescribed paralog, NOTCH2tv. Finally, we apply Fiber-seq and long-read transcript sequencing to human cortical neurospheres to characterize the regulatory landscape and find that the most fixed paralogs, NOTCH2 and NOTCH2NLA, harbor the greatest number of paralog-specific elements potentially driving their regulation.
Keywords: NOTCH2; NOTCH2NL; Segmental duplication; gene duplications; human evolution.
Conflict of interest statement
DECLARATIONS OF INTEREST E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc. A.B.S. is a co-inventor on a patent relating to the Fiber-seq method (US17/995,058). All other authors declare no competing interests.
Figures




References
-
- Yoo DongAhn et al. Complete sequencing of ape genomes. Pages: 2024.07.31.605654 Section:New Results. July 31, 2024. doi: 10.1101/2024.07.31.605654. - DOI
-
- Florio Marta, Heide Michael, et al. “Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex”. In: eLife 7 (Mar. 21, 2018). Ed. by Gleeson Joseph G. Publisher: eLife Sciences Publications, Ltd, e32332. issn: 2050–084X. doi: 10.7554/eLife.32332. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
