This is a preprint.
Intrinsic OASL expression licenses interferon induction during influenza A virus infection
- PMID: 40166309
- PMCID: PMC11956916
- DOI: 10.1101/2025.03.14.643375
Intrinsic OASL expression licenses interferon induction during influenza A virus infection
Abstract
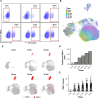
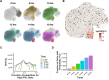
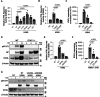
Effective control of viral infection requires rapid induction of the innate immune response, especially the type I and type III interferon (IFN) systems. Despite the critical role of IFN induction in host defense, numerous studies have established that most cells fail to produce IFNs in response to viral stimuli. The specific factors that govern cellular heterogeneity in IFN induction potential during infection are not understood. To identify specific host factors that license some cells but not others to mount an IFN response to viral infection, we developed an approach for analyzing temporal scRNA-seq data of influenza A virus (IAV)-infected cells. This approach identified the expression of several interferon stimulated genes (ISGs) within pre-infection cells as correlates of IFN induction potential of those cells, post-infection. Validation experiments confirmed that intrinsic expression of the ISG OASL is essential for robust IFNL induction during IAV infection. Altogether, our findings reveal an important role for IFN-independent, intrinsic expression of ISGs in promoting IFN induction and provide new insights into the mechanisms that regulate cell-to-cell heterogeneity in innate immune activation.
Keywords: OASL; RNA sensing; RNA velocity; antiviral response; cellular heterogeneity; influenza A virus; innate immunity; interferon; interferon-stimulated genes; scRNA-seq.
Conflict of interest statement
Declaration of interests The authors declare that no competing interests exist.
Figures


Similar articles
-
Fluorescence-Activated Cell Sorting-Based Analysis Reveals an Asymmetric Induction of Interferon-Stimulated Genes in Response to Seasonal Influenza A Virus.J Virol. 2015 Jul;89(14):6982-93. doi: 10.1128/JVI.00857-15. Epub 2015 Apr 22. J Virol. 2015. PMID: 25903337 Free PMC article.
-
Hemagglutinin of Influenza A Virus Antagonizes Type I Interferon (IFN) Responses by Inducing Degradation of Type I IFN Receptor 1.J Virol. 2015 Dec 16;90(5):2403-17. doi: 10.1128/JVI.02749-15. J Virol. 2015. PMID: 26676772 Free PMC article.
-
Casein Kinase 1α Mediates the Degradation of Receptors for Type I and Type II Interferons Caused by Hemagglutinin of Influenza A Virus.J Virol. 2018 Mar 14;92(7):e00006-18. doi: 10.1128/JVI.00006-18. Print 2018 Apr 1. J Virol. 2018. PMID: 29343571 Free PMC article.
-
Innate and adaptive immune responses in HCV infections.J Hepatol. 2014 Nov;61(1 Suppl):S14-25. doi: 10.1016/j.jhep.2014.06.035. Epub 2014 Nov 3. J Hepatol. 2014. PMID: 25443342 Review.
-
The Association of OASL and Type I Interferons in the Pathogenesis and Survival of Intracellular Replicating Bacterial Species.Front Cell Infect Microbiol. 2017 May 19;7:196. doi: 10.3389/fcimb.2017.00196. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28580319 Free PMC article. Review.
References
-
- Galani I.E., Triantafyllia V., Eleminiadou E.-E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., et al. (2017). Interferon-λ Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity 46, 875–890.e6. 10.1016/j.immuni.2017.04.025. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
