This is a preprint.
Proteomic Analysis of Endemic Viral Infections in Neurons offers Insights into Neurodegenerative Diseases
- PMID: 40166347
- PMCID: PMC11957066
- DOI: 10.1101/2025.03.17.643709
Proteomic Analysis of Endemic Viral Infections in Neurons offers Insights into Neurodegenerative Diseases
Abstract
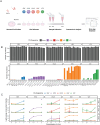
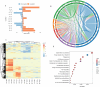
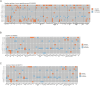
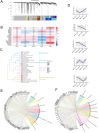
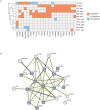
Endemic viral infections with low pathogenicity are often overlooked due to their mild symptoms, yet they can exert long-term effects on cellular function and contribute to disease pathogenesis. While viral infections have been implicated in neurodegenerative disorders, their impact on the neuronal proteome remains poorly understood. Here, we differentiated human induced pluripotent stem cells (KOLF2.1J) into mature neurons to investigate virus-induced proteomic changes following infection with five neurotropic endemic human viruses: Herpes simplex virus 1 (HSV-1), Human coronavirus 229E (HCoV-229E), Epstein-Barr virus (EBV), Varicella-Zoster virus (VZV), and Influenza A virus (H1N1). Given that these viruses can infect adults and have the potential to cross the placental barrier, their molecular impact on neurons may be relevant across the lifespan. Using mass spectrometry-based proteomics with a customized library for simultaneous detection of human and viral proteins, we confirmed successful infections and identified virus-specific proteomic signatures. Notably, virus-induced protein expression changes converged on key neuronal pathways, including those associated with neurodegeneration. Gene co-expression network analysis identified protein modules correlated with viral proteins. Pathway enrichment analysis of these modules revealed associations with the nervous system, including pathways linked to Alzheimer's and Parkinson's disease. Remarkably, several viral-induced proteomic alterations overlapped with changes observed in postmortem Alzheimer's patient brains, suggesting a mechanistic connection between viral exposure and neurodegenerative disease progression. These findings provide molecular insights into how common viral infections perturb neuronal homeostasis and may contribute to neurodegenerative pathology, highlighting the need to consider endemic viruses as potential environmental risk factors in neurological disorders.
Keywords: Alzheimer’s; KOLF2.1J; Neurodegeneration; Neuroinflammation; Neurons; Proteomics; Virus.
Conflict of interest statement
M.A.N., C.W., and Z.L.’s participation in this project was part of a competitive contract awarded to DataTecnica LLC by the National Institutes of Health to support open science research. M.A.N. also currently serves on the scientific advisory board for Character Bio Inc. and is a scientific founder at Neuron23 Inc.
Figures
References
-
- Levine K.S., Leonard H.L., Blauwendraat C., Iwaki H., Johnson N., Bandres-Ciga S., Ferrucci L., Faghri F., Singleton A.B., and Nalls M.A. (2023). Virus exposure and neurodegenerative disease risk across national biobanks. Neuron 111, 1086–1093 e1082. 10.1016/j.neuron.2022.12.029. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
