This is a preprint.
PPM1M, a LRRK2-counteracting, phosphoRab12-preferring phosphatase with potential link to Parkinson's disease
- PMID: 40166354
- PMCID: PMC11957146
- DOI: 10.1101/2025.03.19.644182
PPM1M, a LRRK2-counteracting, phosphoRab12-preferring phosphatase with potential link to Parkinson's disease
Update in
-
PPM1M, an LRRK2-counteracting, phosphoRab12-preferring phosphatase with a potential link to Parkinson's disease.Cell Rep. 2025 Aug 26;44(8):116031. doi: 10.1016/j.celrep.2025.116031. Epub 2025 Jul 20. Cell Rep. 2025. PMID: 40690364 Free PMC article.
Abstract
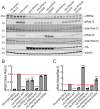
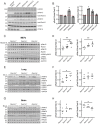
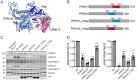
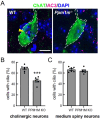
Leucine-rich repeat kinase 2 (LRRK2) phosphorylates a subset of Rab GTPases that regulate receptor trafficking; activating mutations in LRRK2 are linked to Parkinson's disease. Rab phosphorylation is a transient event that can be reversed by phosphatases, including PPM1H, that acts on phosphoRab8A and phosphoRab10. Here we report a phosphatome-wide siRNA screen that identified PPM1M as a phosphoRab12-preferring phosphatase that also acts on phosphoRab8A and phosphoRab10. Upon knockout from cells or mice, PPM1M displays selectivity for phosphoRab12. As shown previously for mice harboring LRRK2 pathway mutations, knockout of Ppm1m leads to primary cilia loss in striatal cholinergic interneurons. We have also identified a rare PPM1M mutation in patients with Parkinson's disease that is catalytically inactive when tested in vitro and in cells. These findings identify PPM1M as a key player in the LRRK2 signaling pathway and provide a new therapeutic target for the possible benefit of patients with Parkinson's disease.
Conflict of interest statement
Competing interests: P.B. and C.B. are employees of CENTOGENE GmbH (Rostock, Germany); all other authors declare that they have no competing interests.
Figures




References
-
- Alessi D. R., Sammler E., LRRK2 kinase in Parkinson’s disease. Science 360, 36–37 (2018). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
