A Smart Single-Loop-Mediated Isothermal Amplification Facilitates Flexible SNP Probe Design for On-Site Rapid Differentiation of SARS-CoV-2 Omicron Variants
- PMID: 40167300
- PMCID: PMC12245020
- DOI: 10.1002/advs.202502708
A Smart Single-Loop-Mediated Isothermal Amplification Facilitates Flexible SNP Probe Design for On-Site Rapid Differentiation of SARS-CoV-2 Omicron Variants
Abstract
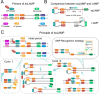
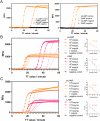
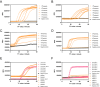
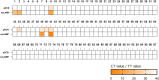
Rapid on-site typing methods for SARS-CoV-2 variants of concern are crucial for its effective surveillance and control. Herein, a smart single-loop-mediated isothermal amplification (ssLAMP) method with the absence of an inner primer but the addition of a swarm primer for differentiation of SARS-CoV-2 Omicron variants is developed. This unique primer design strategy offers greater flexibility in introducing single nucleotide polymorphism (SNP) identification probes and enables multiple detection assays for SARS-CoV-2 Omicron variants including BA.1, BA.2, BA.3, BA.4, and BA.5. A 3D-printed portable dual fluorescence visualization device and smartphone app are developed to enable point-of-care testing. This assay is rapid (within 90 min), highly sensitive (100 copies/reaction), and specific (identification of SNP) for SARA-CoV-2 Omicron variants. The ssLAMP method identifies five BA.5-positive samples among 97 nasopharyngeal swab samples from the clinic, with a 100% concordance rate with Sanger sequencing. The ssLAMP assay system is expected to be utilized for on-site, highly specific, and rapid visualization detection of SARS-CoV-2 and its variants, with great application potential in pathogen genotyping, early cancer screening, and other areas of SNP mutation detection.
Keywords: SARS‐CoV‐2 variants of concern; SNP detection; nucleic acid amplification; ssLAMP.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
An alternative real-time fluorescence reverse transcription loop-mediated isothermal amplification assay for the rapid detection of SARS-CoV-2.Rev Inst Med Trop Sao Paulo. 2025 Jun 27;67:e37. doi: 10.1590/S1678-9946202567037. eCollection 2025. Rev Inst Med Trop Sao Paulo. 2025. PMID: 40608622 Free PMC article.
-
Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa.Lancet Glob Health. 2025 Jul;13(7):e1258-e1267. doi: 10.1016/S2214-109X(25)00150-0. Lancet Glob Health. 2025. PMID: 40580991 Free PMC article.
-
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya.mSphere. 2025 Jun 25;10(6):e0002725. doi: 10.1128/msphere.00027-25. Epub 2025 May 29. mSphere. 2025. PMID: 40439429 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
References
-
- Kim S., Misra A., Annu. Rev. Biomed. Eng. 2007, 9, 289. - PubMed
-
- W. H. O. Coronavirus, (COVID‐19) Dashboard >Cases, [Dashboard] , https://data.who.int/dashboards/covid19/cases (accessed: November 2024).
-
- a) Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., Yu Y., Wang P., Zhang Z., Liu P., An R., Hao X., Wang Y., Wang J., Feng R., Sun H., Zhao L., Zhang W., Zhao D., Zheng J., Yu L., Li C., Zhang N., Wang R., Niu X., Yang S., et al., Nature 2022, 608, 593; - PMC - PubMed
- b) He P., Liu B., Gao X., Yan Q., Pei R., Sun J., Chen Q., Hou R., Li Z., Zhang Y., Zhao J., Sun H., Feng B., Wang Q., Yi H., Hu P., Li P., Zhang Y., Chen Z., Niu X., Zhong X., Jin L., Liu X., Qu K., Ciazynska K. A., Carter A. P., Briggs J. A. G., Chen J., Liu J., Chen X., et al., Nat. Microbiol. 2022, 7, 1635; - PMC - PubMed
- c) Rasmussen A. L., Popescu S. V., Science 2021, 371, 1206. - PubMed
-
- Brito A. F., Semenova E., Dudas G., Hassler G. W., Kalinich C. C., Kraemer M. U. G., Ho J., Tegally H., Githinji G., Agoti C. N., Matkin L. E., Whittaker C., Howden B. P., Sintchenko V., Zuckerman N. S., Mor O., Blankenship H. M., de Oliveira T., Lin R. T. P., Siqueira M. M., Resende P. C., Vasconcelos A. T. R., Spilki F. R., Aguiar R. S., Alexiev I., Ivanov I. N., Philipova I., Carrington C. V. F., Sahadeo N. S. D., Branda B., et al., Nat. Commun. 2022, 13, 7003. - PMC - PubMed
MeSH terms
Supplementary concepts
Grants and funding
- 2022YFD1800400/National Key Research and Development Program of China
- 2023B10564003/Specific university discipline construction project
- R2021PY-QY006/Special Fund for Scientific Innovation Strategy Construction of High Level Academy of Agriculture Science
- 2023QZ-NK13/Opening Project of State Key Laboratory of Swine and Poultry Breeding Industry
- GDNKY-ZQQZ-K25/Opening Project of State Key Laboratory of Swine and Poultry Breeding Industry
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
