Prolonged signaling of backbone-modified glucagon-like peptide- 1 analogues with diverse receptor trafficking
- PMID: 40168114
- PMCID: PMC12002026
- DOI: 10.1073/pnas.2407574122
Prolonged signaling of backbone-modified glucagon-like peptide- 1 analogues with diverse receptor trafficking
Abstract
Signal duration and subcellular location are emerging as important facets of G protein-coupled receptor (GPCR) function. The glucagon-like peptide-1 receptor (GLP-1R), a clinically relevant class B1 GPCR, stimulates production of the second messenger cyclic adenosine monophosphate (cAMP) upon activation by the native hormone, GLP-1. cAMP production continues after the hormone-receptor complex has been internalized via endocytosis. Here, we report GLP-1 analogues that induce prolonged signaling relative to GLP-1. A single β-amino acid substitution at position 18, with the residue derived from (S,S)-trans-2-aminocyclopentanecarboxylic acid (ACPC), enhances signaling duration with retention of receptor endocytosis. Pairing ACPC at position 18 with a second substitution, α-aminoisobutyric acid (Aib) at position 16, abrogates endocytosis, but prolonged signaling is maintained. Prolonged signaling is sensitive to the structure of the β residue at position 18. Cryoelectron microscopy structures of two GLP-1 analogues bound to the GLP-1R:Gs complex suggest substantial alterations to bound peptide structure and dynamics compared to the GLP-1:GLP-1R:Gs complex. These structural findings strengthen an emerging view that agonist dynamics in the receptor-bound state influence signaling profiles. Our results advance understanding of the structural underpinnings of receptor activation and introduce tools for exploring the impact of spatiotemporal signaling profiles following GLP-1R activation.
Keywords: GLP-1; cryo-EM; dynamics; peptide; trafficking.
Conflict of interest statement
Competing interests statement:S.H.G. is a cofounder and shareholder of Longevity Biotech, Inc., which is pursuing biomedical applications of α/β-peptides. P.M.S. is a co-founder and shareholder of Septerna Inc. D.W. is a shareholder of Septerna Inc. P.M.S. and D.W. are co-founders and shareholders of Dacra Therapeutics.
Figures
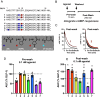
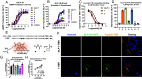

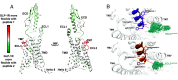
References
-
- Lane J. R., May L. T., Parton R. G., Sexton P. M., Christopoulos A., A kinetic view of GPCR allostery and biased agonism. Nat. Chem. Biol. 13, 929–937 (2017). - PubMed
-
- Grundmann M., Kostenis E., Temporal Bias: Time-encoded dynamic GPCR signaling. Trends Pharmacol. Sci. 38, 1110–1124 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- 1155302/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- T32 GM008505/GM/NIGMS NIH HHS/United States
- IC200100052/Department of Education and Training | Australian Research Council (ARC)
- 2026300/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- R35 GM151985/GM/NIGMS NIH HHS/United States
- R01 GM056414/GM/NIGMS NIH HHS/United States
- 1150083/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- 2025694/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- GM151985/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- DGE-1747503/NSF | NSF Graduate Research Fellowship Program (GRFP)
- T32 GM008349/GM/NIGMS NIH HHS/United States
- 1184726/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- 1154434/Federal Government | DHAC | National Health and Medical Research Council (NHMRC)
- GM056414/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources

