Diverse priming outcomes under conditions of very rare precursor B cells
- PMID: 40168992
- PMCID: PMC12060733
- DOI: 10.1016/j.immuni.2025.03.003
Diverse priming outcomes under conditions of very rare precursor B cells
Abstract
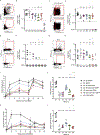
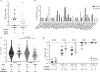
Rare naive B cells have special pathogen-recognition features that enable outsized contributions to protective immunity but infrequently participate in immune responses. We investigatee how germline-targeting vaccine delivery and adjuvant selection affect priming of exceptionally rare BG18-like HIV broadly neutralizing antibody-precursor B cells (<1-in-50 million) in non-human primates. Only escalating dose (ED) priming immunization using the saponin adjuvant SMNP elicited detectable BG18-like cells in germinal centers (GCs) compared with other conditions. All groups had strong GC responses, but only ED+SMNP and bolus+SMNP induced BG18-like memory B cells in >50% of animals. One group had vaccine-specific GC responses equivalent to ED+SMNP but scarce BG18-like B cells. Following homologous boosting, BG18-like memory B cells were present in a bolus priming group but with lower somatic hypermutation and affinities than ED+SMNP. This outcome inversely associated with post-prime antibody titers, suggesting antibody feedback significantly influences rare precursor B cell responses. Thus, antigen and inflammatory stimuli extensively impact priming and affinity maturation of rare B cells.
Keywords: BG18; HIV bnAbs; HIV vaccines; adjuvants; bnAb precursors; escalating dose; germline-targeting; naive B cells.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.J.I. and S.C. are inventors on a patent for the SMNP adjuvant (US11547672B2). K.A.R. and D.J.I. are inventors on patent applications for the synergistic combination of alum and SMNP adjuvants (PCT/US2022/074302 and US No. 17/816,045). D.J.I. and W.R.S. are inventors on a patent for pSer technology (US No. 11,224,648 B2). J.M.S. and W.R.S. are inventors on patent applications related to N332-GT5 that have been filed by Scripps and IAVI. W.R.S. is an employee and shareholder of Moderna, Inc. All other authors declare no competing interests.
Figures
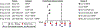




Update of
-
Diverse priming outcomes under conditions of very rare precursor B cells.bioRxiv [Preprint]. 2024 Nov 25:2024.11.21.624746. doi: 10.1101/2024.11.21.624746. bioRxiv. 2024. Update in: Immunity. 2025 Apr 8;58(4):997-1014.e11. doi: 10.1016/j.immuni.2025.03.003. PMID: 39651117 Free PMC article. Updated. Preprint.
References
-
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6. 10.1016/j.immuni.2017.11.023. - DOI - PMC - PubMed
-
- Havenar-Daughton C, Sarkar A, Kulp DW, Toy L, Hu X, Deresa I, Kalyuzhniy O, Kaushik K, Upadhyay AA, Menis S, et al. (2018). The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Science Translational Medicine 10, eaat0381. 10.1126/scitranslmed.aat0381. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

