Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic
- PMID: 40168993
- PMCID: PMC7617805
- DOI: 10.1016/j.cell.2025.02.032
Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic
Abstract
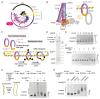
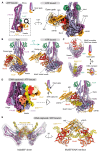
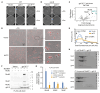
Ring-like structural maintenance of chromosome (SMC) complexes are crucial for genome organization and operate through mechanisms of DNA entrapment and loop extrusion. Here, we explore the DNA loading process of the bacterial SMC complex MukBEF. Using cryoelectron microscopy (cryo-EM), we demonstrate that ATP binding opens one of MukBEF's three potential DNA entry gates, exposing a DNA capture site that positions DNA at the open neck gate. We discover that the gp5.9 protein of bacteriophage T7 blocks this capture site by DNA mimicry, thereby preventing DNA loading and inactivating MukBEF. We propose a comprehensive and unidirectional loading mechanism in which DNA is first captured at the complex's periphery and then ingested through the DNA entry gate, powered by a single cycle of ATP hydrolysis. These findings illuminate a fundamental aspect of how ubiquitous DNA organizers are primed for genome maintenance and demonstrate how this process can be disrupted by viruses.
Keywords: DNA mimics; MukBEF; SMC complexes; Wadjet; bacteriophages; cohesin; condensin; cryo-EM.
Copyright © 2025 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Yatskevich S, Rhodes J, Nasmyth K. Organization of chromosomal DNA by SMC complexes. Annu Rev Genet. 2019;53:445–482. - PubMed
-
- Gruber S. SMC complexes sweeping through the chromosome: going with the flow and against the tide. Curr Opin Microbiol. 2018;42:96–103. - PubMed
-
- Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17:1014–1023. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

