Sexual selection, genomic evolution and population fitness in Drosophila pseudoobscura
- PMID: 40169023
- PMCID: PMC11961267
- DOI: 10.1098/rspb.2024.2744
Sexual selection, genomic evolution and population fitness in Drosophila pseudoobscura
Abstract
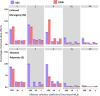
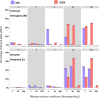
Sexual selection shapes the genome in unique ways. It is also likely to have significant fitness consequences, such as purging deleterious mutations from the genome or conversely maintaining genetic load in a population via sexual conflict. Here, we examined what the influence of sexual selection has on genomic variation potentially underlying population fitness using experimentally evolved Drosophila pseudoobscura populations. Sexual selection was manipulated by keeping replicate lines in elevated polyandry or strict monogamy for approximately 200 generations followed by individual-based sequencing. Using pi (π), fixation index (Fst)and recombination rate measures, we confirmed signatures of selection were not dispersed but mainly localized to the third and X chromosome. Overall mutational load was similar between lines but our analysis of the distribution of fitness effects revealed considerable variation between lines and chromosomes. Furthermore, we found that the distribution of transposable elements differs between the lines, with a higher load in monogamous lines. Our results suggest that complex interactions between purifying selection and sexual conflict are shaping the genome, particularly on chromosome 3 and the sex chromosome; sexual selection influences divergence across chromosomes but in a more complex way than proposed by simple 'purging' of deleterious loci.
Keywords: distribution of fitness effects; mutational load; sexual conflict; sexual selection.
Conflict of interest statement
We declare we have no competing interests.
Figures




References
-
- Maggu K, Kapse S, Ahlawat N, Geeta Arun M, Prasad NG. 2022. Finding love: fruit fly males evolving under higher sexual selection are inherently better at finding receptive females. Anim. Behav. 187, 15–33. (10.1016/j.anbehav.2022.02.010) - DOI
-
- Clutton-Brock T. 2009. Sexual selection in females. Anim. Behav. 77, 3–11. (10.1016/j.anbehav.2008.08.026) - DOI
-
- Karsten KB, Andriamandimbiarisoa LN, Fox SF, Raxworthy CJ. 2009. Sexual selection on body size and secondary sexual characters in 2 closely related, sympatric chameleons in Madagascar. Behav. Ecol. 20, 1079–1088. (10.1093/beheco/arp100) - DOI
-
- Jaiswal SK, Gupta A, Shafer ABA, Prasoodanan P K V, Vijay N, Sharma VK. 2021. Genomic insights into the molecular basis of sexual selection in birds. Front. Ecol. Evol. 9. (10.3389/fevo.2021.538498) - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

