Transcriptomic and spatial GABAergic neuron subtypes in zona incerta mediate distinct innate behaviors
- PMID: 40169544
- PMCID: PMC11961626
- DOI: 10.1038/s41467-025-57896-2
Transcriptomic and spatial GABAergic neuron subtypes in zona incerta mediate distinct innate behaviors
Abstract
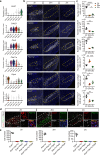
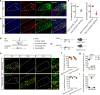
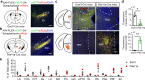
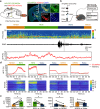
Understanding the anatomical connection and behaviors of transcriptomic neuron subtypes is critical to delineating cell type-specific functions in the brain. Here we integrated single-nucleus transcriptomic sequencing, in vivo circuit mapping, optogenetic and chemogenetic approaches to dissect the molecular identity and function of heterogeneous GABAergic neuron populations in the zona incerta (ZI) in mice, a region involved in modulating various behaviors. By microdissecting ZI for transcriptomic and spatial gene expression analyses, our results revealed two non-overlapping Ecel1- and Pde11a-expressing GABAergic neurons with dominant expression in the rostral and medial zona incerta (ZIrEcel1 and ZImPde11a), respectively. The GABAergic projection from ZIrEcel1 to periaqueductal gray mediates self-grooming, while the GABAergic projection from ZImPde11a to the oral part of pontine reticular formation promotes transition from sleep to wakefulness. Together, our results revealed the molecular markers, spatial organization and specific neuronal circuits of two discrete GABAergic projection neuron populations in segregated subregions of the ZI that mediate distinct innate behaviors, advancing our understanding of the functional organization of the brain.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Zeng, H. & Sanes, J. R. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci.18, 530–546 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

