Severe inflammation and lineage skewing are associated with poor engraftment of engineered hematopoietic stem cells in patients with sickle cell disease
- PMID: 40169559
- PMCID: PMC11961595
- DOI: 10.1038/s41467-025-58321-4
Severe inflammation and lineage skewing are associated with poor engraftment of engineered hematopoietic stem cells in patients with sickle cell disease
Abstract
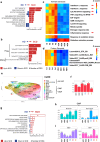
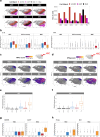
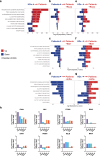
In sickle cell disease (SCD), the β6Glu→Val substitution in the β-globin leads to red blood cell sickling. The transplantation of autologous, genetically modified hematopoietic stem and progenitor cells (HSPCs) is a promising treatment option for patients with SCD. We completed a Phase I/II open-label clinical trial (NCT03964792) for patients with SCD using a lentiviral vector (DREPAGLOBE) expressing a potent anti-sickling β-globin. The primary endpoint was to evaluate the short-term safety and secondary endpoints included the efficacy and the long-term safety. We report on the results after 18 to 36 months of follow-up. No drug-related adverse events or signs of clonal hematopoiesis were observed. Despite similar vector copy numbers in the drug product, gene-marking in peripheral blood mononuclear cells and correction of the clinical phenotype varied from one patient to another. Single-cell transcriptome analyses show that in the patients with poor engraftment, the most immature HSCs display an exacerbated inflammatory signature (via IL-1 or TNF-α and interferon signaling pathways). This signature is accompanied by a lineage bias in the HSCs. Our clinical data indicates that the DREPAGLOBE-based gene therapy (GT) is safe. However, its efficacy is variable and probably depends on the number of infused HSCs and intrinsic, engraftment-impairing inflammatory alterations in HSCs. Trial: NCT03964792.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Marktel, S. et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ß-thalassemia. Nat. Med.25, 234–241 (2019). - PubMed
-
- Levasseur, D. N. et al. A recombinant human hemoglobin with anti-sickling properties greater than fetal hemoglobin. J. Biol. Chem.279, 27518–27524 (2004). - PubMed
-
- Ribeil, J.-A. et al. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med.376, 848–855 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

