Somatic NAP1L1 p.D349E promotes cardiac hypertrophy through cGAS-STING-IFN signaling
- PMID: 40169585
- PMCID: PMC11961713
- DOI: 10.1038/s41467-025-58453-7
Somatic NAP1L1 p.D349E promotes cardiac hypertrophy through cGAS-STING-IFN signaling
Abstract
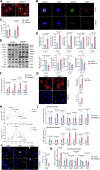
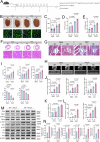
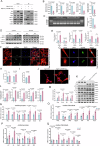
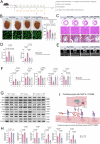
Hypertrophic cardiomyopathy (HCM) is the most common inherited heart disease, often caused by sarcomere gene mutations, though many sporadic cases remain genetically unexplained. Here we show that the somatic variant NAP1L1 p.D349E was involved in cardiac hypertrophy in sporadic HCM patients. Through next generation sequencing, we found that somatic variant NAP1L1 p.D349E was recurrent in the cardiomyocytes of gene-elusive sporadic HCM patients. Subsequent in vivo and in vitro functional analysis confirmed that NAP1L1 p.D349E contributes to HCM by triggering an innate immunity response. This mutation destabilizes nucleosome formation, causing DNA to leak into the cytoplasm. This leakage activates a key immune pathway, cGAS-STING, which leads to the release of inflammatory molecules and promotes heart muscle thickening. Our findings reveal a new mechanism driving HCM and suggest that somatic variants could be important in understanding and management of HCM.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Ommen, S. R. et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J. Am. Coll. Cardiol.76, e159–e240 (2020). - PubMed
-
- Ingles, J. et al. Nonfamilial hypertrophic cardiomyopathy: prevalence, natural history, and clinical implications. Circ. Cardiovasc. Genet. 10, 10.1161/CIRCGENETICS.116.001620 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

